Transcription
Sakshi Education
Transcription is the process by which genetic information from DNA is transferred into RNA. In simple terms DNA dependent RNA synthesis is called transcription. The process of transcription is accomplished by an enzymatic system consisting of RNA polymerase and other accessory or auxillary proteins. The stretch of DNA that is transcribed into an RNA molecule is called a transcription unit.
In case of prokaryotic transcription occurs in the cytoplasm simultaneously with translation. Therefore transcription and translation are a coupled process. On the other hand, transcription in eukaryotes is primarily localized to the nucleus and is separated from the cytoplasm by the nuclear membrane. The complimentary RNA transcript formed is then transported into the cytoplasm where translation occurs. Therefore, transcription and translation are spatially and temporally separated.
Comparision of Transcription and Replication:
 s factor is responsible for promoter specificity and is not a part of the core enzyme. The RNA polymerase core enzyme can bind to DNA but cannot initiate transcription on its own and requires the aid of sigma factor s. s factor alone cannot bind to the DNA as its DNA binding site is blocked and opens only when it binds to the RNA polymerase core enzyme.
s factor is responsible for promoter specificity and is not a part of the core enzyme. The RNA polymerase core enzyme can bind to DNA but cannot initiate transcription on its own and requires the aid of sigma factor s. s factor alone cannot bind to the DNA as its DNA binding site is blocked and opens only when it binds to the RNA polymerase core enzyme.
Different sigma factors recognize different promoters but can bind to the same core polymerase. Sigma factor are synthesized at different time points and thus affect the expression of genes differentially forming a means for differential gene expression.
The product of transcription is called as primary transcript which is nascent RNA molecule. The primary transcript after undergoing modifications is called as secondary transcript or matured transcript. Unlike DNA polymerase RNA polymerase does not have proof reading activity (3’-5’ exonuclease).
In case of prokaryotes a single RNA polymerase is responsible for synthesis of all RNA molecules.3 types of RNA molecules are formed by transcription
Transcriptional unit in case of eukaryotes generally contains the information for only one gene. In case of proaryotes one transcriptional unit may contain information for more than one gene.
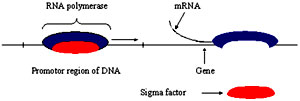
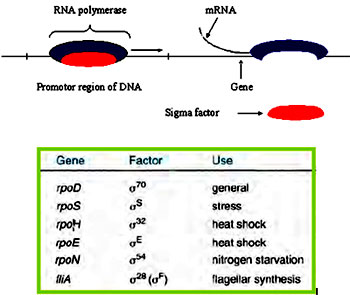
Promoter: The region of DNA where RNA polymerase binds and accomplishes transcription
 Recognition of promoter regions: Recognition of promoter is crucial for transcription initiation and is the first step of gene expression and is the point where majority of the gene regulation mechanisms are exercised. RNA polymerase core enzyme can bind to any region of DNA non specifically by means of general affinity. This type of binding is called loose binding. Initiation of transcription is facilitated at the promoter regions but the core enzyme cannot differentiate between the promoter regions and any other DNA regions. Affinity of the core enzyme for the promoter regions is altered by the binding of sigma factor. Sigma factor increases the affinity of the core enzyme for the promoters by 1000-10,000 times. The site where the first nucleotide in the RNA is complimentarily base paired to the DNA sequence is regarded as the Transcriptional start site (TS) and it is denoted as +1. The region of DNA present towards the 5’ end of the start site is considered as the Upstream region and is denoted with negative integers such as -1.-2.-3 etc. Similarly, region of DNA present towards the 3’ end of the start site is considered as the Downstream region and is denoted with positive integers such as +1.+2.+3 etc.
Recognition of promoter regions: Recognition of promoter is crucial for transcription initiation and is the first step of gene expression and is the point where majority of the gene regulation mechanisms are exercised. RNA polymerase core enzyme can bind to any region of DNA non specifically by means of general affinity. This type of binding is called loose binding. Initiation of transcription is facilitated at the promoter regions but the core enzyme cannot differentiate between the promoter regions and any other DNA regions. Affinity of the core enzyme for the promoter regions is altered by the binding of sigma factor. Sigma factor increases the affinity of the core enzyme for the promoters by 1000-10,000 times. The site where the first nucleotide in the RNA is complimentarily base paired to the DNA sequence is regarded as the Transcriptional start site (TS) and it is denoted as +1. The region of DNA present towards the 5’ end of the start site is considered as the Upstream region and is denoted with negative integers such as -1.-2.-3 etc. Similarly, region of DNA present towards the 3’ end of the start site is considered as the Downstream region and is denoted with positive integers such as +1.+2.+3 etc.
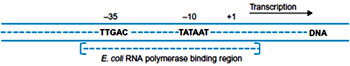
The RNA polymerase holoenzyme recognizes the promoters specifically by identifying consensus sequences present at -10 and -35 regions and binds to it. This is called Tight binding and the polymerase bound to double stranded DNA complex is called as Closed binary complex. In case of prokaryotes at the consensus sequence present at -10 region is “TATAAT” and the region is called “TATA box”. TATA box in prokaryotes is called as Pribnow box.
The consensus sequence present at -35 region is “TAGACA”.
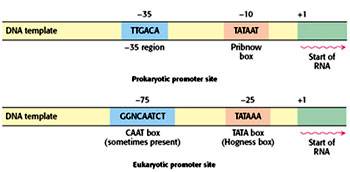 The -35 region is essential for recognition of the promoters by the RNA polymerase holoenzyme and the -10 region is essential for promoter melting. The distance between -10 and -35 region for optimum gene expression is 17-19 base pairs. Any mutation at -10 region or at -35 region that alters the sequences in those regions making it more dissimilar will decrease the degree of gene expressions. Any mutation in the spacer regions between -10 and -35 regions that either increase or decrease the distance between the two regions may also decrease the degree of gene expression. Such mutations are called as Down mutations. Mutations that increase the degree of gene expression are called Up-mutations. Mutations in -10 regions are found to be more deleterious than mutations in -35 region. Mutations in the spacer region that alters the phasing of the consensus sequences such that both the sequences are present on opposite sites are more deleterious than mutations that bring both the sequences onto the same phase. For instance insertional mutations of five nucleotides are more deleterious than insertion of ten nucleotides.
The -35 region is essential for recognition of the promoters by the RNA polymerase holoenzyme and the -10 region is essential for promoter melting. The distance between -10 and -35 region for optimum gene expression is 17-19 base pairs. Any mutation at -10 region or at -35 region that alters the sequences in those regions making it more dissimilar will decrease the degree of gene expressions. Any mutation in the spacer regions between -10 and -35 regions that either increase or decrease the distance between the two regions may also decrease the degree of gene expression. Such mutations are called as Down mutations. Mutations that increase the degree of gene expression are called Up-mutations. Mutations in -10 regions are found to be more deleterious than mutations in -35 region. Mutations in the spacer region that alters the phasing of the consensus sequences such that both the sequences are present on opposite sites are more deleterious than mutations that bring both the sequences onto the same phase. For instance insertional mutations of five nucleotides are more deleterious than insertion of ten nucleotides.
Sigma factor alone cannot bind to the promoter region because of the presence of an N-terminal autoinhibitary domain. It blocks the DNA binding site of the sigma factor When sigma factor combines with RNA core enzyme sigma factor undergo conformational changes such that the DNA binding site can bind to the promoters.
 Eukaryotic genes encoding proteins have promoter sites with a TATAAA consensus sequence, called a TATA box or a Hogness box, centered at about –25. Many eukaryotic promoters also have a CAAT box with a GGNCAATCT consensus sequence centered at about –75. Transcription of eukaryotic genes is further stimulated by enhancer sequences, which can be quite distant (as many as several kilo bases) from the start site, on either its 5’ or its 3’ side.
Eukaryotic genes encoding proteins have promoter sites with a TATAAA consensus sequence, called a TATA box or a Hogness box, centered at about –25. Many eukaryotic promoters also have a CAAT box with a GGNCAATCT consensus sequence centered at about –75. Transcription of eukaryotic genes is further stimulated by enhancer sequences, which can be quite distant (as many as several kilo bases) from the start site, on either its 5’ or its 3’ side.
Transcription initiation
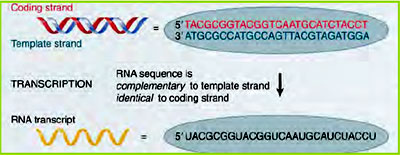
In transcription, one strand of DNA, the non-coding strand, is used as a template for RNA synthesis. As transcription proceeds in the 5' ? 3' direction, and uses base pairing complimentarity with the DNA template to specify the correct copying, the DNA template strand is that oriented in the 3' ? 5' direction. The strand that is not used as the template is called the coding strand, and has the DNA sequence that reflects that of the RNA produced.
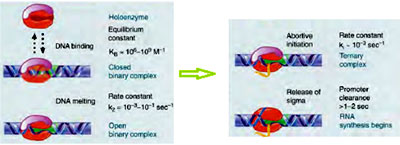 Once the RNA polymerase holoenzyme recognizes the promoters, it binds and initiates the transcription The RNA polymerase core enzyme associated with a sigma factor 70 (the general transcription factor) that aids in finding the appropriate -35 and -10 base pairs in the promoter sequences. Sigma factor is essential for unwinding the promoter region – Promoter Melting. Upon melting, the single stranded DNA obtained is used as template for synthesizing the RNA. This complex of melted DNA associated with the polymerase is called Open Binary Complex (DNA being one component and Polymerase the other component). The initial nucleotides of RNA bind to the template DNA by complimentary base pairing which involves only a few hydrogen bonds. The energy involved in these hydrogen bonds is not sufficient enough for the RNA molecule to bind to the template for longer time because of this the RNA molecule detaches from the template DNA and the transcription process is stopped. This is called Abortive initiation. RNA polymerase holoenzyme makes multiple attempts to synthesisze the RNA molecule in a continuous manner
Once the RNA polymerase holoenzyme recognizes the promoters, it binds and initiates the transcription The RNA polymerase core enzyme associated with a sigma factor 70 (the general transcription factor) that aids in finding the appropriate -35 and -10 base pairs in the promoter sequences. Sigma factor is essential for unwinding the promoter region – Promoter Melting. Upon melting, the single stranded DNA obtained is used as template for synthesizing the RNA. This complex of melted DNA associated with the polymerase is called Open Binary Complex (DNA being one component and Polymerase the other component). The initial nucleotides of RNA bind to the template DNA by complimentary base pairing which involves only a few hydrogen bonds. The energy involved in these hydrogen bonds is not sufficient enough for the RNA molecule to bind to the template for longer time because of this the RNA molecule detaches from the template DNA and the transcription process is stopped. This is called Abortive initiation. RNA polymerase holoenzyme makes multiple attempts to synthesisze the RNA molecule in a continuous manner
Once RNA polymerase adds about 9 to 10 nucleotides the DNA-RNA hybrid becomes stable and the RNA polymerase moves forward to add further nucleotides. This phenomenon is called as Promoter clearance. Promoter clearance is an indication of successful transcription initiation
After transcription initiation open binary complex is converted into an open ternary complex.
RNA polymerase transcribes the DNA synthesising RNA in 5’-3’ direction. Ribonucleotide triphosphates act as substrates for transcription which are converted in ribonucleotide monophosphates in the RNA molecule. RNA polymerase does not have 3’-5’ exonuclease activity and hence cannot carry out proof reading. The rate of transcription is approximately 40 nt/sec.
Eukaryotic Transcription Initiation:
 Transcription in eukaryotes is carried out by atleast three different types of RNA polymerases which recognizes different of promoters and therefore genes, Interesting feature of these polymerases is that they exhibit different differential sensitivity to the toxin, a-amanitin (obtained from Amanita philloides) Transcription initiation is far more complex in eukaryotes and the main difference being that eukaryotic polymerases do not recognize directly their core promoter sequences.
Transcription in eukaryotes is carried out by atleast three different types of RNA polymerases which recognizes different of promoters and therefore genes, Interesting feature of these polymerases is that they exhibit different differential sensitivity to the toxin, a-amanitin (obtained from Amanita philloides) Transcription initiation is far more complex in eukaryotes and the main difference being that eukaryotic polymerases do not recognize directly their core promoter sequences.
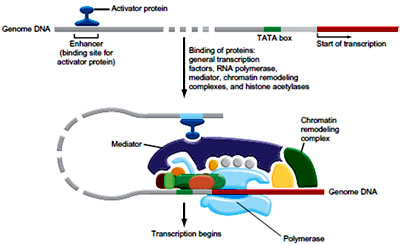
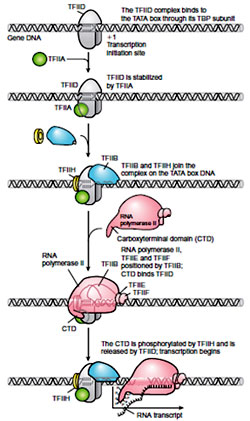 As mentioned above recognition of promoters involves the identification of the TATA box. In case of the eukaryotes the task is performed by a protein called the transcription factor IID (TFIID) and more precisely by one of the subunits of TFIID known as the TATA-binding protein, or TBP. Several other transcription factors (TFIIA, TFIIB, TFIIE, TFIIF and TFIIH) then bind to TFIID and to the promoter region. TFIIF is the protein that guides RNA polymerase II to the beginning of the gene to be transcribed. The complex formed between the TATA box, TFIID, the other transcritpion factors, and RNA polymerase is known as the transcription preinitiation complex. Note that the proteins with the prefix TFII are so named because they are transcription factors that help RNA polymerase II to bind to promoter sequences.
As mentioned above recognition of promoters involves the identification of the TATA box. In case of the eukaryotes the task is performed by a protein called the transcription factor IID (TFIID) and more precisely by one of the subunits of TFIID known as the TATA-binding protein, or TBP. Several other transcription factors (TFIIA, TFIIB, TFIIE, TFIIF and TFIIH) then bind to TFIID and to the promoter region. TFIIF is the protein that guides RNA polymerase II to the beginning of the gene to be transcribed. The complex formed between the TATA box, TFIID, the other transcritpion factors, and RNA polymerase is known as the transcription preinitiation complex. Note that the proteins with the prefix TFII are so named because they are transcription factors that help RNA polymerase II to bind to promoter sequences.
Eukaryotic RNA polymerases and effect of a-amanitin:
Transcription elongation: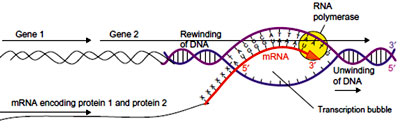 After transcription initiation sigma factor may be released. The RNA polymerase core enzyme moves forward unwinding the double helix to expose new segment of the template in single stranded condition
After transcription initiation sigma factor may be released. The RNA polymerase core enzyme moves forward unwinding the double helix to expose new segment of the template in single stranded condition
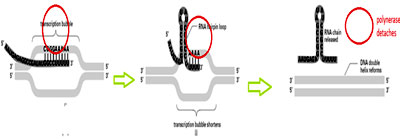
The enzyme adds nucleotides to the 3’ end of the growing RNA during this process the ß and ? phosphates from the incoming NTPs are removed and remaining NMPs are added onto the 3’OH group of the existing ribonucleotide. The RNA- DNA hybrid formed during transcription assumes A- form of double helix during transcription. As the enzyme moves forward it unwinds the region in front and at the same time single stranded DNA is renatured at the rear end. In this manner, a transcription bubble of length 12-14 nucleotides is maintained during the elongation process.
Termination:
There are two basic mechanisms of transcription termination IN prokaryotes..
 The signal for transcriptional termination is present in the secondary structure of the RNA. In case of intrinsic termination the termination signal sequences present in the RNA are enough for transcription termination and do not require the help of Rho protein. The signal for transcription termination is in the form of GC rich region followed by an AT rich region. GC rich region is present in the form of inverted repeats which is complimentary to each other. As the RNA polymerase transcribes the termination region the GC rich region forms a hairpin structure in the RNA molecule. RNA polymerase has more affinity for double stranded RNA than for DNA The ß subunit of RNA polymerase binds to the hairpin structure present behind the polymerase. Because of the binding of RNA polymerase to the hairpin structure it leads to Pausing of the RNA polymerase. At this stage if the RNA polymerase finds a continuous stretch of adenines in the template it further slows down as the probability of finding the same nucleotides by the RNA pol will be less. In addition, as the dA:dU interactions are the weakest interactions, the RNA polymerase loses its affinity and detaches from the template DNA releasing the nascent RNA molecule.
The signal for transcriptional termination is present in the secondary structure of the RNA. In case of intrinsic termination the termination signal sequences present in the RNA are enough for transcription termination and do not require the help of Rho protein. The signal for transcription termination is in the form of GC rich region followed by an AT rich region. GC rich region is present in the form of inverted repeats which is complimentary to each other. As the RNA polymerase transcribes the termination region the GC rich region forms a hairpin structure in the RNA molecule. RNA polymerase has more affinity for double stranded RNA than for DNA The ß subunit of RNA polymerase binds to the hairpin structure present behind the polymerase. Because of the binding of RNA polymerase to the hairpin structure it leads to Pausing of the RNA polymerase. At this stage if the RNA polymerase finds a continuous stretch of adenines in the template it further slows down as the probability of finding the same nucleotides by the RNA pol will be less. In addition, as the dA:dU interactions are the weakest interactions, the RNA polymerase loses its affinity and detaches from the template DNA releasing the nascent RNA molecule.
Extrinsic termination or Rho dependent Termination:
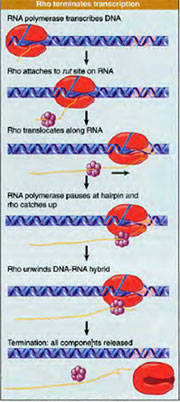 In case of some genes termination signals are weak and require the help of an external part for transcription termination. Rho protein is a helicase which is involved in extrinsic termination. In extrinsic terminators the GC rich region is not immediately followed by AT rich region. AT rich region is present much farther away or is absent because of this during transcription RNA polymerase pauses but does not detach from the template. Rho protein binds to the RNA transcript and moves towards 3’end of the RNA i.e growing end when the RNA polymerase pauses the Rho protein reaches the transcription bubble and exhibits helicase activity. As a consequence, the RNA transcript is released and the RNA polymerase detaches from the template. The region where the transcription process stops releasing the RNA molecule because of the detachment of RNA polymerase from the template is called as termination region.
In case of some genes termination signals are weak and require the help of an external part for transcription termination. Rho protein is a helicase which is involved in extrinsic termination. In extrinsic terminators the GC rich region is not immediately followed by AT rich region. AT rich region is present much farther away or is absent because of this during transcription RNA polymerase pauses but does not detach from the template. Rho protein binds to the RNA transcript and moves towards 3’end of the RNA i.e growing end when the RNA polymerase pauses the Rho protein reaches the transcription bubble and exhibits helicase activity. As a consequence, the RNA transcript is released and the RNA polymerase detaches from the template. The region where the transcription process stops releasing the RNA molecule because of the detachment of RNA polymerase from the template is called as termination region.
Inhibition of Transcription:
In case of prokaryotic transcription occurs in the cytoplasm simultaneously with translation. Therefore transcription and translation are a coupled process. On the other hand, transcription in eukaryotes is primarily localized to the nucleus and is separated from the cytoplasm by the nuclear membrane. The complimentary RNA transcript formed is then transported into the cytoplasm where translation occurs. Therefore, transcription and translation are spatially and temporally separated.
Comparision of Transcription and Replication:
- The process of transcription is similar to DNA replication in many ways, as both the processes are template dependent. For DNA replication the DNA polymerase binds to single stranded DNA template. In transcription RNA polymerase binds to double stranded DNA but subsequently uses single stranded DNA as a template.
- Both replication and transcription occurs in 5’-3’ direction. The reaction mechanism is same for both.
- The process of polymerisation is a SN2 nucelophillic substitution reaction., where the 3’OH group of the sugar of the existing nucleotide makes a nucleophilic attack on the a-Phosphate group of the incoming nucleotide tri phosphate (NTP).
- The principle involved in both the processes is complimentary base pairing to the existing template.
- DNA synthesis involves Deoxy ribonucleotides and transcription involves ribonucleotides, where the substrates for replication of DNA are dNTPs - dATP, dGTP, dCTP, dTTP and for transcription NTPs are substrates- ATP, GTP, dCTP and UTP.
- DNA replication initiation requires primers whereas transcription does not require any primers.
- The first nucleotide in the RNA molecule is in the form of NTP whereas in case of DNA it is in the form of nucleotide mono phosphate (NMP).( This enables the RNA to undergo 5’capping to protect it from nucleotide degradation, and is not possible in DNA )
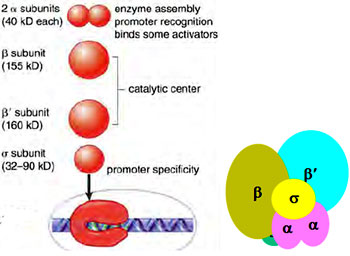
- a- subunits form the basement upon which the ß subunits attach and another important feature of the a- subunits is that they interact with activator proteins. a- subunits also provide binding site for s factor.
- ß and ß’ form the active site which catalyses the formation of phosphodiester bond.
- ? subunit is not functionally characterized.
- a2 , ß, ß’, ? forms core polymerase.
- a2 , ß, ß’, ? along with s factor forms the holoenzyme.
 s factor is responsible for promoter specificity and is not a part of the core enzyme. The RNA polymerase core enzyme can bind to DNA but cannot initiate transcription on its own and requires the aid of sigma factor s. s factor alone cannot bind to the DNA as its DNA binding site is blocked and opens only when it binds to the RNA polymerase core enzyme.
s factor is responsible for promoter specificity and is not a part of the core enzyme. The RNA polymerase core enzyme can bind to DNA but cannot initiate transcription on its own and requires the aid of sigma factor s. s factor alone cannot bind to the DNA as its DNA binding site is blocked and opens only when it binds to the RNA polymerase core enzyme.
Different sigma factors recognize different promoters but can bind to the same core polymerase. Sigma factor are synthesized at different time points and thus affect the expression of genes differentially forming a means for differential gene expression.
The product of transcription is called as primary transcript which is nascent RNA molecule. The primary transcript after undergoing modifications is called as secondary transcript or matured transcript. Unlike DNA polymerase RNA polymerase does not have proof reading activity (3’-5’ exonuclease).
In case of prokaryotes a single RNA polymerase is responsible for synthesis of all RNA molecules.3 types of RNA molecules are formed by transcription
- r-RNA
- t-RNA
- m-RNA
- r-RNA: It is ribosomal RNA as it becomes an integrsl part of ribosomes. Therefore ribosomes are called as Ribonucleoproteins. They doesnot undergo translation.There are 3 r-RNA molecules in prokaryotes -23S, 16S and 5S.
- t-RNA (transfer RNA): is involved in the transfer of amino acids to the site of protein synthesis and is also called as soluble RNA or adaptor RNA.
- m-RNA (messenger RNA): This RNA molecule is involved in the transfer of genetic information in the RNA molecule into proteins. It acts as template for translation.
- microRNA (abbreviated miRNA): is a small non coding molecule of about 22 nucleotides found in plants, animals, and some viruses, which functions in transcriptional and post-transcriptional regulation of gene expression.
- sn RNA (Small nuclear RNA): These are small RNA molecules which are non coding and are present in the nucleus. These RNA molecules are involved in the regulation of gene expression.
- Hn-RNA (Heterogenous nuclear RNA): It is a mixture of RNA molecules existing in different stages of processing in the nucleus of eukaryotes.
Transcriptional unit in case of eukaryotes generally contains the information for only one gene. In case of proaryotes one transcriptional unit may contain information for more than one gene.
Promoter: The region of DNA where RNA polymerase binds and accomplishes transcription
- Promoter are A-T rich sequences that promote transcription by providing single stranded DNA template, as such can undergo denaturation easily
- Promoters are usually upstream but they can be present within the coding sequences also ( downstream stream or internal promoters)
- Promoters and other DNA binding sequences can be identified by DNA Footprinting technique or Enzyme Mobility Shift Assay (EMSAis also known as Gel retardation assay)
- Recognition of promoter regions
- Transcriptional initiation
- Transcriptional elongation
- Transcriptional termination
 Recognition of promoter regions: Recognition of promoter is crucial for transcription initiation and is the first step of gene expression and is the point where majority of the gene regulation mechanisms are exercised. RNA polymerase core enzyme can bind to any region of DNA non specifically by means of general affinity. This type of binding is called loose binding. Initiation of transcription is facilitated at the promoter regions but the core enzyme cannot differentiate between the promoter regions and any other DNA regions. Affinity of the core enzyme for the promoter regions is altered by the binding of sigma factor. Sigma factor increases the affinity of the core enzyme for the promoters by 1000-10,000 times. The site where the first nucleotide in the RNA is complimentarily base paired to the DNA sequence is regarded as the Transcriptional start site (TS) and it is denoted as +1. The region of DNA present towards the 5’ end of the start site is considered as the Upstream region and is denoted with negative integers such as -1.-2.-3 etc. Similarly, region of DNA present towards the 3’ end of the start site is considered as the Downstream region and is denoted with positive integers such as +1.+2.+3 etc.
Recognition of promoter regions: Recognition of promoter is crucial for transcription initiation and is the first step of gene expression and is the point where majority of the gene regulation mechanisms are exercised. RNA polymerase core enzyme can bind to any region of DNA non specifically by means of general affinity. This type of binding is called loose binding. Initiation of transcription is facilitated at the promoter regions but the core enzyme cannot differentiate between the promoter regions and any other DNA regions. Affinity of the core enzyme for the promoter regions is altered by the binding of sigma factor. Sigma factor increases the affinity of the core enzyme for the promoters by 1000-10,000 times. The site where the first nucleotide in the RNA is complimentarily base paired to the DNA sequence is regarded as the Transcriptional start site (TS) and it is denoted as +1. The region of DNA present towards the 5’ end of the start site is considered as the Upstream region and is denoted with negative integers such as -1.-2.-3 etc. Similarly, region of DNA present towards the 3’ end of the start site is considered as the Downstream region and is denoted with positive integers such as +1.+2.+3 etc.The RNA polymerase holoenzyme recognizes the promoters specifically by identifying consensus sequences present at -10 and -35 regions and binds to it. This is called Tight binding and the polymerase bound to double stranded DNA complex is called as Closed binary complex. In case of prokaryotes at the consensus sequence present at -10 region is “TATAAT” and the region is called “TATA box”. TATA box in prokaryotes is called as Pribnow box.
The consensus sequence present at -35 region is “TAGACA”.
 The -35 region is essential for recognition of the promoters by the RNA polymerase holoenzyme and the -10 region is essential for promoter melting. The distance between -10 and -35 region for optimum gene expression is 17-19 base pairs. Any mutation at -10 region or at -35 region that alters the sequences in those regions making it more dissimilar will decrease the degree of gene expressions. Any mutation in the spacer regions between -10 and -35 regions that either increase or decrease the distance between the two regions may also decrease the degree of gene expression. Such mutations are called as Down mutations. Mutations that increase the degree of gene expression are called Up-mutations. Mutations in -10 regions are found to be more deleterious than mutations in -35 region. Mutations in the spacer region that alters the phasing of the consensus sequences such that both the sequences are present on opposite sites are more deleterious than mutations that bring both the sequences onto the same phase. For instance insertional mutations of five nucleotides are more deleterious than insertion of ten nucleotides.
The -35 region is essential for recognition of the promoters by the RNA polymerase holoenzyme and the -10 region is essential for promoter melting. The distance between -10 and -35 region for optimum gene expression is 17-19 base pairs. Any mutation at -10 region or at -35 region that alters the sequences in those regions making it more dissimilar will decrease the degree of gene expressions. Any mutation in the spacer regions between -10 and -35 regions that either increase or decrease the distance between the two regions may also decrease the degree of gene expression. Such mutations are called as Down mutations. Mutations that increase the degree of gene expression are called Up-mutations. Mutations in -10 regions are found to be more deleterious than mutations in -35 region. Mutations in the spacer region that alters the phasing of the consensus sequences such that both the sequences are present on opposite sites are more deleterious than mutations that bring both the sequences onto the same phase. For instance insertional mutations of five nucleotides are more deleterious than insertion of ten nucleotides.Sigma factor alone cannot bind to the promoter region because of the presence of an N-terminal autoinhibitary domain. It blocks the DNA binding site of the sigma factor When sigma factor combines with RNA core enzyme sigma factor undergo conformational changes such that the DNA binding site can bind to the promoters.
 Eukaryotic genes encoding proteins have promoter sites with a TATAAA consensus sequence, called a TATA box or a Hogness box, centered at about –25. Many eukaryotic promoters also have a CAAT box with a GGNCAATCT consensus sequence centered at about –75. Transcription of eukaryotic genes is further stimulated by enhancer sequences, which can be quite distant (as many as several kilo bases) from the start site, on either its 5’ or its 3’ side.
Eukaryotic genes encoding proteins have promoter sites with a TATAAA consensus sequence, called a TATA box or a Hogness box, centered at about –25. Many eukaryotic promoters also have a CAAT box with a GGNCAATCT consensus sequence centered at about –75. Transcription of eukaryotic genes is further stimulated by enhancer sequences, which can be quite distant (as many as several kilo bases) from the start site, on either its 5’ or its 3’ side. Transcription initiation
In transcription, one strand of DNA, the non-coding strand, is used as a template for RNA synthesis. As transcription proceeds in the 5' ? 3' direction, and uses base pairing complimentarity with the DNA template to specify the correct copying, the DNA template strand is that oriented in the 3' ? 5' direction. The strand that is not used as the template is called the coding strand, and has the DNA sequence that reflects that of the RNA produced.
 Once the RNA polymerase holoenzyme recognizes the promoters, it binds and initiates the transcription The RNA polymerase core enzyme associated with a sigma factor 70 (the general transcription factor) that aids in finding the appropriate -35 and -10 base pairs in the promoter sequences. Sigma factor is essential for unwinding the promoter region – Promoter Melting. Upon melting, the single stranded DNA obtained is used as template for synthesizing the RNA. This complex of melted DNA associated with the polymerase is called Open Binary Complex (DNA being one component and Polymerase the other component). The initial nucleotides of RNA bind to the template DNA by complimentary base pairing which involves only a few hydrogen bonds. The energy involved in these hydrogen bonds is not sufficient enough for the RNA molecule to bind to the template for longer time because of this the RNA molecule detaches from the template DNA and the transcription process is stopped. This is called Abortive initiation. RNA polymerase holoenzyme makes multiple attempts to synthesisze the RNA molecule in a continuous manner
Once the RNA polymerase holoenzyme recognizes the promoters, it binds and initiates the transcription The RNA polymerase core enzyme associated with a sigma factor 70 (the general transcription factor) that aids in finding the appropriate -35 and -10 base pairs in the promoter sequences. Sigma factor is essential for unwinding the promoter region – Promoter Melting. Upon melting, the single stranded DNA obtained is used as template for synthesizing the RNA. This complex of melted DNA associated with the polymerase is called Open Binary Complex (DNA being one component and Polymerase the other component). The initial nucleotides of RNA bind to the template DNA by complimentary base pairing which involves only a few hydrogen bonds. The energy involved in these hydrogen bonds is not sufficient enough for the RNA molecule to bind to the template for longer time because of this the RNA molecule detaches from the template DNA and the transcription process is stopped. This is called Abortive initiation. RNA polymerase holoenzyme makes multiple attempts to synthesisze the RNA molecule in a continuous mannerOnce RNA polymerase adds about 9 to 10 nucleotides the DNA-RNA hybrid becomes stable and the RNA polymerase moves forward to add further nucleotides. This phenomenon is called as Promoter clearance. Promoter clearance is an indication of successful transcription initiation
After transcription initiation open binary complex is converted into an open ternary complex.
RNA polymerase transcribes the DNA synthesising RNA in 5’-3’ direction. Ribonucleotide triphosphates act as substrates for transcription which are converted in ribonucleotide monophosphates in the RNA molecule. RNA polymerase does not have 3’-5’ exonuclease activity and hence cannot carry out proof reading. The rate of transcription is approximately 40 nt/sec.
Eukaryotic Transcription Initiation:
 Transcription in eukaryotes is carried out by atleast three different types of RNA polymerases which recognizes different of promoters and therefore genes, Interesting feature of these polymerases is that they exhibit different differential sensitivity to the toxin, a-amanitin (obtained from Amanita philloides) Transcription initiation is far more complex in eukaryotes and the main difference being that eukaryotic polymerases do not recognize directly their core promoter sequences.
Transcription in eukaryotes is carried out by atleast three different types of RNA polymerases which recognizes different of promoters and therefore genes, Interesting feature of these polymerases is that they exhibit different differential sensitivity to the toxin, a-amanitin (obtained from Amanita philloides) Transcription initiation is far more complex in eukaryotes and the main difference being that eukaryotic polymerases do not recognize directly their core promoter sequences.  As mentioned above recognition of promoters involves the identification of the TATA box. In case of the eukaryotes the task is performed by a protein called the transcription factor IID (TFIID) and more precisely by one of the subunits of TFIID known as the TATA-binding protein, or TBP. Several other transcription factors (TFIIA, TFIIB, TFIIE, TFIIF and TFIIH) then bind to TFIID and to the promoter region. TFIIF is the protein that guides RNA polymerase II to the beginning of the gene to be transcribed. The complex formed between the TATA box, TFIID, the other transcritpion factors, and RNA polymerase is known as the transcription preinitiation complex. Note that the proteins with the prefix TFII are so named because they are transcription factors that help RNA polymerase II to bind to promoter sequences.
As mentioned above recognition of promoters involves the identification of the TATA box. In case of the eukaryotes the task is performed by a protein called the transcription factor IID (TFIID) and more precisely by one of the subunits of TFIID known as the TATA-binding protein, or TBP. Several other transcription factors (TFIIA, TFIIB, TFIIE, TFIIF and TFIIH) then bind to TFIID and to the promoter region. TFIIF is the protein that guides RNA polymerase II to the beginning of the gene to be transcribed. The complex formed between the TATA box, TFIID, the other transcritpion factors, and RNA polymerase is known as the transcription preinitiation complex. Note that the proteins with the prefix TFII are so named because they are transcription factors that help RNA polymerase II to bind to promoter sequences. Eukaryotic RNA polymerases and effect of a-amanitin:
| Type | Location | Cellular Transcripts | Effects of a-amanitin |
| I | Nucleolus | 18S, 5.8S and 28S rRNA | Insensitive |
| II | Nucleoplasm | mRNA precursors, most snRNA, miRNA, siRNA, snoRNA | Strongly inhibited even at low concentrations |
| III | Nucleoplasm | tRNA and 5S rRNA, some snRNA and other ncRNAs | Inhibited by high concentrations |
Transcription elongation:
 After transcription initiation sigma factor may be released. The RNA polymerase core enzyme moves forward unwinding the double helix to expose new segment of the template in single stranded condition
After transcription initiation sigma factor may be released. The RNA polymerase core enzyme moves forward unwinding the double helix to expose new segment of the template in single stranded conditionThe enzyme adds nucleotides to the 3’ end of the growing RNA during this process the ß and ? phosphates from the incoming NTPs are removed and remaining NMPs are added onto the 3’OH group of the existing ribonucleotide. The RNA- DNA hybrid formed during transcription assumes A- form of double helix during transcription. As the enzyme moves forward it unwinds the region in front and at the same time single stranded DNA is renatured at the rear end. In this manner, a transcription bubble of length 12-14 nucleotides is maintained during the elongation process.
Termination:
There are two basic mechanisms of transcription termination IN prokaryotes..
- Intrinsic or Rho independent
- Extrinsic or Rho dependent
 The signal for transcriptional termination is present in the secondary structure of the RNA. In case of intrinsic termination the termination signal sequences present in the RNA are enough for transcription termination and do not require the help of Rho protein. The signal for transcription termination is in the form of GC rich region followed by an AT rich region. GC rich region is present in the form of inverted repeats which is complimentary to each other. As the RNA polymerase transcribes the termination region the GC rich region forms a hairpin structure in the RNA molecule. RNA polymerase has more affinity for double stranded RNA than for DNA The ß subunit of RNA polymerase binds to the hairpin structure present behind the polymerase. Because of the binding of RNA polymerase to the hairpin structure it leads to Pausing of the RNA polymerase. At this stage if the RNA polymerase finds a continuous stretch of adenines in the template it further slows down as the probability of finding the same nucleotides by the RNA pol will be less. In addition, as the dA:dU interactions are the weakest interactions, the RNA polymerase loses its affinity and detaches from the template DNA releasing the nascent RNA molecule.
The signal for transcriptional termination is present in the secondary structure of the RNA. In case of intrinsic termination the termination signal sequences present in the RNA are enough for transcription termination and do not require the help of Rho protein. The signal for transcription termination is in the form of GC rich region followed by an AT rich region. GC rich region is present in the form of inverted repeats which is complimentary to each other. As the RNA polymerase transcribes the termination region the GC rich region forms a hairpin structure in the RNA molecule. RNA polymerase has more affinity for double stranded RNA than for DNA The ß subunit of RNA polymerase binds to the hairpin structure present behind the polymerase. Because of the binding of RNA polymerase to the hairpin structure it leads to Pausing of the RNA polymerase. At this stage if the RNA polymerase finds a continuous stretch of adenines in the template it further slows down as the probability of finding the same nucleotides by the RNA pol will be less. In addition, as the dA:dU interactions are the weakest interactions, the RNA polymerase loses its affinity and detaches from the template DNA releasing the nascent RNA molecule.Extrinsic termination or Rho dependent Termination:
 In case of some genes termination signals are weak and require the help of an external part for transcription termination. Rho protein is a helicase which is involved in extrinsic termination. In extrinsic terminators the GC rich region is not immediately followed by AT rich region. AT rich region is present much farther away or is absent because of this during transcription RNA polymerase pauses but does not detach from the template. Rho protein binds to the RNA transcript and moves towards 3’end of the RNA i.e growing end when the RNA polymerase pauses the Rho protein reaches the transcription bubble and exhibits helicase activity. As a consequence, the RNA transcript is released and the RNA polymerase detaches from the template. The region where the transcription process stops releasing the RNA molecule because of the detachment of RNA polymerase from the template is called as termination region.
In case of some genes termination signals are weak and require the help of an external part for transcription termination. Rho protein is a helicase which is involved in extrinsic termination. In extrinsic terminators the GC rich region is not immediately followed by AT rich region. AT rich region is present much farther away or is absent because of this during transcription RNA polymerase pauses but does not detach from the template. Rho protein binds to the RNA transcript and moves towards 3’end of the RNA i.e growing end when the RNA polymerase pauses the Rho protein reaches the transcription bubble and exhibits helicase activity. As a consequence, the RNA transcript is released and the RNA polymerase detaches from the template. The region where the transcription process stops releasing the RNA molecule because of the detachment of RNA polymerase from the template is called as termination region.Inhibition of Transcription:
- Rifampicin is a semi synthetic derivative of rifamycins, which are derived from strain of Streptomyces. It interferes with the formation of the first few phosphodiester bonds in the RNA chain.
- Actinomycin D, a polypeptide containing antibiotic from a different strain of Streptomyces. It bind tightly and specifically to double-helical DNA and thereby prevents it form being an effective template for RNA synthesis.
Published date : 20 Jun 2014 05:03PM






