Nucleic Acids- Discovery and Structure
Sakshi Education
Discovery of Nucleic Acids - Friedrich Miescher, 1869
Miescher was the first person who isolated the nuclei of white blood cells, obtained from pus cells. His experiments revealed that nuclei contained a chemical that contained nitrogen and phosphorus but no sulfur. He called the chemical nuclein because it came from nuclei and since they exhibited acidic nature (as they were negatively charged )it was later termed as nucleic acid.
Proteins Produce Genetic Traits - Archibald Garrod, 1909
Garrod noticed that people with certain genetic abnormalities (inborn errors of metabolism) lacked certain enzymes. This observation linked proteins (enzymes) to genetic traits.
Genetic Material can Transform Bacteria - Frederick Griffith, 1931
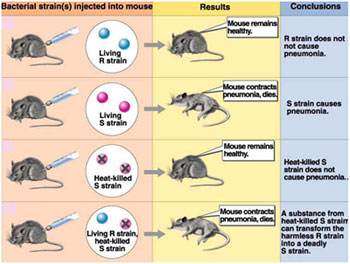
In 1928, Frederick Griffith was trying to find a vaccine which could offer protection against the disease- pneumonia, caused by the bacterium, Streptococcus pneumoniae. He isolated two strains of the bacterium. One had a polysaccharide capsule that gave it a smooth (S) appearance when observed for colony morphology in culture. The other form appeared rough (R) in culture as it lacked the capsule. The ‘S’ form acts as the virulent form of the bacterium, since the capsule protects it from getting recognized by the immune system of the host and other harmful things in its environment.
Griffith injected these bacteria into mice, and observed the consequence. Interestingly, he found mice injected with S forms died, whereas the mice injected with R forms were alive. Mice injected with heat-killed S forms lived. But: Mice injected with a mixture of heat-killed S forms and live R forms died, and when isolated, contained live S form bacteria.
What did this mean?
Today, transformation is defined as the process by which external DNA is assimilated into a cell changing its genotype and phenotype. Transformation is one of the processes used in DNA technologies.
The transforming material is DNA - Oswald Avery, Colin MacLeod, and Maclyn McCarty, 1944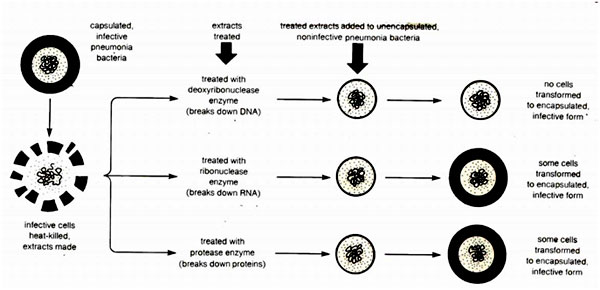
Structural Evidences Supporting DNA as the Genetic Molecule
From this information, James Watson and, Francis Crick (who died in July, 2004 at the age of 88) determined the structure of DNA in 1953 and published their work in Nature. They solved the structure of DNA and gave the following postulates:
DNA exists in numerous possible confirmations depending on the nucleotide sequence of the DNA, the degree and direction of supercoiling, chemical modifications of the bases and also solution conditions, such as the concentration of metal ions used as counter ions. The conformations so far identified are: A-DNA, B-DNA, C-DNA, D-DNA, E-DNA, H-DNA, L-DNA, and Z-DNA. However, only A-DNA, B-DNA, and Z-DNA are believed to be found in nature. Amongst these three conformations, the "B" form is most common under the conditions found in cells. The two alternative double-helical forms of DNA differ in their geometry and dimensions.
B-DNA:
DNA assumes the so called B-confirmation indicated by their X-ray diffraction patterns, when it is isolated using counter ions which are alkali metals such as Na+ and the relative humidity is >92%. B-DNA is regarded as the native confirmation (biologically functional) form of DNA and the same can be noticed from the x-ray diffraction pattern of the DNA in intact sperm heads. B-DNA consist of two polynucleotide chains that wind about a common axis with a right handed twist to form a 20°A diameter helix.
Two strands are antiparallel and wrap around each other such that they cannot be separated without unwinding the helix and such type of coiling is known as Pleotropic coiling. The planes of the bases are nearly perpendicular to the helix axis. Two bases forms H-bonds and make planar base pairing. The ideal B-DNA helix has 10bp per turn (360°) a helical twist of 36° per base pair and since the aromatic bases have vanderwaals thickness of 3.4A° (0.34nm) and are particularly stacked on each other bases (base stacking). A, T and G, C base pairs are interchangeable. B-DNA has two deep exterior grooves that wind between its sugars phosphate chains. Grooves are formed as consequence of geometry of base pairs. The angle at which the two sugars protrude from each base pair the angle between two glycosidic bond is 120° and 240° for wide range. As a consequence, more base pairs stack on top of each other. The narrow angle between the sugars on one edge of the base pair generate minor groove and angle on other edge gives major groove.
IF the sugar points away in straight line i.e. at an angle of 180° then the two grooves would be in equal diameter. Asymmetry of deoxyriobose sugar and the nucleotides are responsible for generation of grooves in DNA helix.
Double helical DNA and RNA assume several distinct structures which vary with factors such as the humidity, the identity of the cations used and also with base sequences.
A-DNA:
When the relative humidity is decreased to 75%. B-DNA undergoes a reversible conformational change, so called A- form. X-ray studies indicate that A-DNA forms a wider and flutter right handed helix than B-DNA, with 11 bp/ turn, a rise per base pair of 2.6 A° and pitch of the helix being 28.6 A°. In A-DNA the plane of the bases is tilted 20° with respect to the helix axis, since its helix passes above the major groove side of the base pairs rather than through them as in B-DNA. The major deep and minor groove is shallow. Most self-complimentary oligonucleotides of <10bp crystallized is the A-DNA confirmation. Like B-DNA these molecules exhibit considerable sequence specific confirmation variation. A-DNA has so far been observed in only three biological contexts.
Z-DNA: (Left handed helix)
The crystal structure determination of the self-complementary hexa nucleotides by Andrew Wang and Alexander Rich surprisingly revealed a left handed double helix. It has 12 Watson crick base pairs per turn, with a pitch of 44A° and in contrast to A-DNA a deep minor groove and no recognizable major groove.
The base pairs in Z-DNA are flipped 180° relative to those in B-DNA, the repeating unit of Z-DNA is dinucleotide. The line joining successive phosphorus atoms on a polynucleotide structure of Z-DNA, therefore follows a zig- zag path around the helix rather than a smooth curve.
X-ray diffraction studies and NMR studies have shown that complementary polynucleotides with alternating purines and pyrimidine such as poly d (GC) and poly d(GC) and poly d (AC), poly d(GT) take up Z-DNA confirmation at high salt concentration. High salt concentration stabilizes electric repulsion between closet approaching phosphate groups on opposite strands.
 Methylation of cytosine at C5 position also promotes Z-DNA formation as the hydrophobic CH3 at this position is less exposed to solvent in Z-DNA than in B-DNA. Reversible conversion of specific segments of B-DNA to Z-DNA under appropriate circumstances acts as a monitoring mechanism in regulating gene expression. There are indicators that it transiently forms behind actively transcribing RNA polymerase but it has been difficult to prove the in vivo existence of Z-DNA due to lack of particular probes for detecting Z-DNA such as Z-DNA specific antibody.
Methylation of cytosine at C5 position also promotes Z-DNA formation as the hydrophobic CH3 at this position is less exposed to solvent in Z-DNA than in B-DNA. Reversible conversion of specific segments of B-DNA to Z-DNA under appropriate circumstances acts as a monitoring mechanism in regulating gene expression. There are indicators that it transiently forms behind actively transcribing RNA polymerase but it has been difficult to prove the in vivo existence of Z-DNA due to lack of particular probes for detecting Z-DNA such as Z-DNA specific antibody.
Miescher was the first person who isolated the nuclei of white blood cells, obtained from pus cells. His experiments revealed that nuclei contained a chemical that contained nitrogen and phosphorus but no sulfur. He called the chemical nuclein because it came from nuclei and since they exhibited acidic nature (as they were negatively charged )it was later termed as nucleic acid.
Proteins Produce Genetic Traits - Archibald Garrod, 1909
Garrod noticed that people with certain genetic abnormalities (inborn errors of metabolism) lacked certain enzymes. This observation linked proteins (enzymes) to genetic traits.
Genetic Material can Transform Bacteria - Frederick Griffith, 1931
In 1928, Frederick Griffith was trying to find a vaccine which could offer protection against the disease- pneumonia, caused by the bacterium, Streptococcus pneumoniae. He isolated two strains of the bacterium. One had a polysaccharide capsule that gave it a smooth (S) appearance when observed for colony morphology in culture. The other form appeared rough (R) in culture as it lacked the capsule. The ‘S’ form acts as the virulent form of the bacterium, since the capsule protects it from getting recognized by the immune system of the host and other harmful things in its environment.
Griffith injected these bacteria into mice, and observed the consequence. Interestingly, he found mice injected with S forms died, whereas the mice injected with R forms were alive. Mice injected with heat-killed S forms lived. But: Mice injected with a mixture of heat-killed S forms and live R forms died, and when isolated, contained live S form bacteria.
What did this mean?
- The production of a capsule is an inheritable trait that distinguishes the R form from the S form of the bacterium.
- Somehow, the heat which killed the S cells did not damage the material that had, genetic instructions, so that this material (instructions on how to make a capsule) could be incorporated into the living R cells (The R cells could pick up this material from the environment) transforming these R cells into virulent S forms.
- Griffith called this the Transformation Principle.
- A back mutation the gene encoding for the capsule restoring its normal phenotype can convert a R strain into a S strain.
- As presumed, above it could be due to transforming principle
Today, transformation is defined as the process by which external DNA is assimilated into a cell changing its genotype and phenotype. Transformation is one of the processes used in DNA technologies.
The transforming material is DNA - Oswald Avery, Colin MacLeod, and Maclyn McCarty, 1944

- Mice + DNA-digesting enzyme + heat-killed S + R ----> Live Mice
- Mice + RNA-digesting enzyme + heat-killed S + R ----> Dead Mice
- Mice + Protein-digesting enzyme + heat-killed S + R --> Dead Mice
In 1944, Avery concluded that the genetic molecule is DNA as the process of transformation was prevented only when DNA was destroyed. Many scientists still disputed this conclusion, since the structure of DNA was not known, and sophisticated protein purification techniques were not available. Many argued that the DNA digesting enzymes which were proteins in nature had other contaminated proteins which were acting as genetic material but not DNA as such. Avery could not disprove the argument with substantial experimental evidence.
From this experiment the important implication drawn was that, Avery, MacLeod, and McCarty proposed for the first time that DNA acts as the genetic material.
More Evidence: The Genetic Material is DNA - Alfred D. Hershey and Martha Chase, 1952
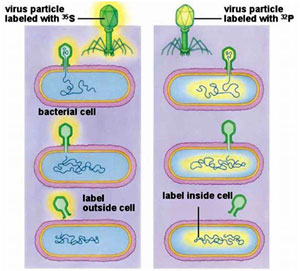 Bacteriophages (viruses that invade bacteria and convert the bacteria into virus making machines) proved to be the means by which, the question was finally answered. In 1952, Hershey and Chase (and others) confirmed that DNA was the genetic molecule. Viruses have just DNA (or sometimes just RNA) and a protein coat. Proteins contain sulfur, but not phosphorus and DNA contains phosphorus, but not sulfur.
Bacteriophages (viruses that invade bacteria and convert the bacteria into virus making machines) proved to be the means by which, the question was finally answered. In 1952, Hershey and Chase (and others) confirmed that DNA was the genetic molecule. Viruses have just DNA (or sometimes just RNA) and a protein coat. Proteins contain sulfur, but not phosphorus and DNA contains phosphorus, but not sulfur.
Hershey and Chase used radioactive Sulfur and Phosphorus to "label" T2 phages. They then tracked the invasion of phages into host bacteria (a strain of E coli) and what part of the new generation phages became radioactive. Since only the DNA of the new generation of phages was radioactive, Hershey and Chase were able to confirm that DNA was the genetic molecule.
Hershey and Chase worked with viruses that infect bacteria called bacteriophages. T phages are temperate phages and act as coliphages (the phages that infect E.coli). The bacteriophage gets adsorbed onto the bacterial sueface, drills a hole through which its genetic material is pushed into the bacterial cell. The viral genetic material gets integrated into the bacterial genome after which the bacterial cell treats the viral genetic material as if it was its own and subsequently manufactures more virus particles. Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria.
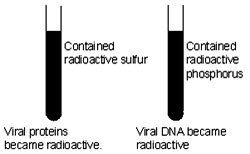 They grew a virus population in medium that contained radioactive phosphorus and another in medium that contained radioactive sulfur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly, viruses grown on radioactive sulfur contained radioactive protein, but not radioactive DNA, because DNA does not contain sulfur.
They grew a virus population in medium that contained radioactive phosphorus and another in medium that contained radioactive sulfur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly, viruses grown on radioactive sulfur contained radioactive protein, but not radioactive DNA, because DNA does not contain sulfur.
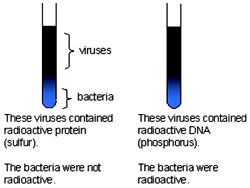 Radioactive bacteriophages were incubated with the E. coli bacteria to facilitate attachment. Then as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. Since, centrifuge was not available in those days and kitchen blendor was used for the purpose, this experiment is also famously known as “Kitchen Blendor “ experiment.
Radioactive bacteriophages were incubated with the E. coli bacteria to facilitate attachment. Then as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. Since, centrifuge was not available in those days and kitchen blendor was used for the purpose, this experiment is also famously known as “Kitchen Blendor “ experiment.
After agitating, the culture was pelleted and the pellet was observed for the radioactivity. Bacteria that were infected with viruses, that had radioactive DNA, were radioactive, indicating that DNA was the material that passed from the virus to the bacteria. Bacteria that were infected with viruses, that had radioactive proteins were not radioactive. This indicates that proteins did not enter the bacteria from the viruses. DNA is therefore the genetic material that is passed from virus to bacteria.
Reconstitution experiments by Singer and Conrat- Discovery of RNA as genetic material.
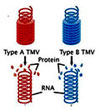 Singer and Conrat performed reconstitution experiments using the Tobacco Mosaic Virus (TMV.) These viruses are classified into different types based type of capsomeres they possess. For instance Type A virus contains Type A capsomeres and Type B virus contains type B capsomeres.
Singer and Conrat performed reconstitution experiments using the Tobacco Mosaic Virus (TMV.) These viruses are classified into different types based type of capsomeres they possess. For instance Type A virus contains Type A capsomeres and Type B virus contains type B capsomeres.
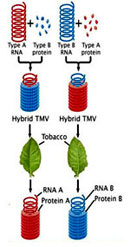 Another important feature of the viruses is that they can be packaged in vitro. When the capsomeres and the genetic material are put together, they self assemble into viral particles. Using this feature, Singer and Conrat, reconstituted hybrid viral particles, that is, a virus containing type A genetic material was mixed with type B capsomeres. Similarly, a virus containing type B genetic material was mixed with type A capsomeres. When these hybrid viruses were infected onto tobacco leaves and the progeny was analyzed, it was surprising to find that type A genetic material produced only type A capsomeres and similarly viruses that had type B genetic material produced only type B.
Another important feature of the viruses is that they can be packaged in vitro. When the capsomeres and the genetic material are put together, they self assemble into viral particles. Using this feature, Singer and Conrat, reconstituted hybrid viral particles, that is, a virus containing type A genetic material was mixed with type B capsomeres. Similarly, a virus containing type B genetic material was mixed with type A capsomeres. When these hybrid viruses were infected onto tobacco leaves and the progeny was analyzed, it was surprising to find that type A genetic material produced only type A capsomeres and similarly viruses that had type B genetic material produced only type B.
From the results of the experiments, it was clearly proved that the nucleic acids propogated form one generation to the other but the proteins did not .Two important conclusions were drawn fron the experiment:\
From this experiment the important implication drawn was that, Avery, MacLeod, and McCarty proposed for the first time that DNA acts as the genetic material.
More Evidence: The Genetic Material is DNA - Alfred D. Hershey and Martha Chase, 1952
 Bacteriophages (viruses that invade bacteria and convert the bacteria into virus making machines) proved to be the means by which, the question was finally answered. In 1952, Hershey and Chase (and others) confirmed that DNA was the genetic molecule. Viruses have just DNA (or sometimes just RNA) and a protein coat. Proteins contain sulfur, but not phosphorus and DNA contains phosphorus, but not sulfur.
Bacteriophages (viruses that invade bacteria and convert the bacteria into virus making machines) proved to be the means by which, the question was finally answered. In 1952, Hershey and Chase (and others) confirmed that DNA was the genetic molecule. Viruses have just DNA (or sometimes just RNA) and a protein coat. Proteins contain sulfur, but not phosphorus and DNA contains phosphorus, but not sulfur.Hershey and Chase used radioactive Sulfur and Phosphorus to "label" T2 phages. They then tracked the invasion of phages into host bacteria (a strain of E coli) and what part of the new generation phages became radioactive. Since only the DNA of the new generation of phages was radioactive, Hershey and Chase were able to confirm that DNA was the genetic molecule.
Hershey and Chase worked with viruses that infect bacteria called bacteriophages. T phages are temperate phages and act as coliphages (the phages that infect E.coli). The bacteriophage gets adsorbed onto the bacterial sueface, drills a hole through which its genetic material is pushed into the bacterial cell. The viral genetic material gets integrated into the bacterial genome after which the bacterial cell treats the viral genetic material as if it was its own and subsequently manufactures more virus particles. Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria.
 They grew a virus population in medium that contained radioactive phosphorus and another in medium that contained radioactive sulfur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly, viruses grown on radioactive sulfur contained radioactive protein, but not radioactive DNA, because DNA does not contain sulfur.
They grew a virus population in medium that contained radioactive phosphorus and another in medium that contained radioactive sulfur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly, viruses grown on radioactive sulfur contained radioactive protein, but not radioactive DNA, because DNA does not contain sulfur.  Radioactive bacteriophages were incubated with the E. coli bacteria to facilitate attachment. Then as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. Since, centrifuge was not available in those days and kitchen blendor was used for the purpose, this experiment is also famously known as “Kitchen Blendor “ experiment.
Radioactive bacteriophages were incubated with the E. coli bacteria to facilitate attachment. Then as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. Since, centrifuge was not available in those days and kitchen blendor was used for the purpose, this experiment is also famously known as “Kitchen Blendor “ experiment. After agitating, the culture was pelleted and the pellet was observed for the radioactivity. Bacteria that were infected with viruses, that had radioactive DNA, were radioactive, indicating that DNA was the material that passed from the virus to the bacteria. Bacteria that were infected with viruses, that had radioactive proteins were not radioactive. This indicates that proteins did not enter the bacteria from the viruses. DNA is therefore the genetic material that is passed from virus to bacteria.
Reconstitution experiments by Singer and Conrat- Discovery of RNA as genetic material.
 Singer and Conrat performed reconstitution experiments using the Tobacco Mosaic Virus (TMV.) These viruses are classified into different types based type of capsomeres they possess. For instance Type A virus contains Type A capsomeres and Type B virus contains type B capsomeres.
Singer and Conrat performed reconstitution experiments using the Tobacco Mosaic Virus (TMV.) These viruses are classified into different types based type of capsomeres they possess. For instance Type A virus contains Type A capsomeres and Type B virus contains type B capsomeres. Another important feature of the viruses is that they can be packaged in vitro. When the capsomeres and the genetic material are put together, they self assemble into viral particles. Using this feature, Singer and Conrat, reconstituted hybrid viral particles, that is, a virus containing type A genetic material was mixed with type B capsomeres. Similarly, a virus containing type B genetic material was mixed with type A capsomeres. When these hybrid viruses were infected onto tobacco leaves and the progeny was analyzed, it was surprising to find that type A genetic material produced only type A capsomeres and similarly viruses that had type B genetic material produced only type B.
Another important feature of the viruses is that they can be packaged in vitro. When the capsomeres and the genetic material are put together, they self assemble into viral particles. Using this feature, Singer and Conrat, reconstituted hybrid viral particles, that is, a virus containing type A genetic material was mixed with type B capsomeres. Similarly, a virus containing type B genetic material was mixed with type A capsomeres. When these hybrid viruses were infected onto tobacco leaves and the progeny was analyzed, it was surprising to find that type A genetic material produced only type A capsomeres and similarly viruses that had type B genetic material produced only type B.From the results of the experiments, it was clearly proved that the nucleic acids propogated form one generation to the other but the proteins did not .Two important conclusions were drawn fron the experiment:\
- TMV has RNA as genetic material.
- RNA acts as Genetic material.
Structural Evidences Supporting DNA as the Genetic Molecule
- Mirsky restated work from the 1850's, that determined the relationship of the volume of DNA in the nucleus for normal body cells,and cells just prior to division and in gametes. This provided evidence that DNA was the genetic molecule because it corresponded with the behavior of chromosomes in mitosis and meiosis.
- Erwin Chargaff's work in 1947:
Erwin Chargaff in 1940, conducted experimental studies that revealed that in DNA the content of adenine and thymine (A=T) is equal and similarly the content of guanine and cytosine (G=C) is also equal. He found that the base composition of DNA from given organism is characteristic of that organism. I.e. it is independent of
- tissues from which the DNA is taken
- organisms age
- nutritional state or any other environment factors
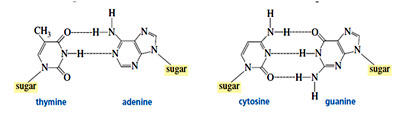
[Adenine] = [Thymine]
[Guanine] = [Cytosine]
[Adenine] + [Guanine] = [Thymine] + [Cytosine]
This rule is not applicable for single stranded nucleic acids. SS DNA viral genetic material when enters it host organism and replicates to for double stranded molecules obeys this rule. Some DNA molecules contain bases that are chemical derivatives of the standard set. The modified bases obeys Chargaff rule, if the derivatives bases are taken as equivalent to their parent bases. Likewise many bases in RNA (particularly t RNA) are derivatives of bases.
- X-ray diffraction studies of DNA, done by Rosalind Franklin and Maurice Wilikins at King's College in London) showed that DNA:
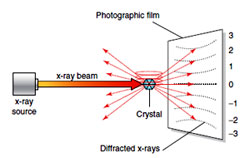
- Was long and thin
- Had a uniform diameter of 2 nanometers
- Had a highly repetitive structure with 0.34 nm between nitrogen bases in the stack
- Was probably helical in shape, like a circular stairway
From this information, James Watson and, Francis Crick (who died in July, 2004 at the age of 88) determined the structure of DNA in 1953 and published their work in Nature. They solved the structure of DNA and gave the following postulates:

- DNA is double stranded in nature. This postulate was important in the context that ruled out the three strand model proposed by Pauling.
- DNA is helical in shape (as determined by X ray diffraction studies of Wilkins and Franklin).
- Two strands are anti parallel in orientation. One strand is oriented in the 5' to 3' direction while the other strand runs in the 3' to 5' direction. Because the two strands are oppositely oriented, they are said to be anti parallel to each other. (This postulate was putforth keeping in mind a model to explain DNA replication).
- In the helical structure, nitrogen base groups lie towards the inside of the helix while the sugar and phosphate groups face outward. The sugar and phosphate groups in the helix therefore make up the backbone of DNA.
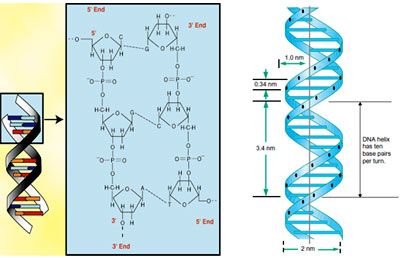
- Nitrogen bases lie perpendicularly to the direction of the helix and can be represented as the steps of the helical ladder while the adjoining rods can be compared to that of the sugar phosphate backbone.
- In the helix the two strands are joined by means of complementary base pairing. Adenine bonds with thymine and cytosine bonds with guanine. The nitrogen bases are held together by hydrogen bonds: adenine and thymine pair with two hydrogen bonds whereas cytosine and guanine pair with three hydrogen bonds. (Based on the Chargaff’s studies)
- The width of the DNA helix is 2 nm which is just sufficient to accommodate a purine and a pyrimidine. (as determined by X ray diffraction studies of Wilkins and Franklin).
- Each turn of the DNA helix takes occupies 34 A? .
- The helix accommodates 10 bp per turn (360°) with a helical twist of 36° per base pair. Each base pair occupies 3.4 A?.
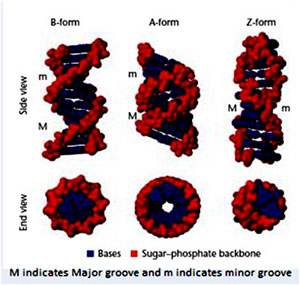
- During the attainment of helical structure, two types of grooves are formed- major grooves and minor grooves.
DNA exists in numerous possible confirmations depending on the nucleotide sequence of the DNA, the degree and direction of supercoiling, chemical modifications of the bases and also solution conditions, such as the concentration of metal ions used as counter ions. The conformations so far identified are: A-DNA, B-DNA, C-DNA, D-DNA, E-DNA, H-DNA, L-DNA, and Z-DNA. However, only A-DNA, B-DNA, and Z-DNA are believed to be found in nature. Amongst these three conformations, the "B" form is most common under the conditions found in cells. The two alternative double-helical forms of DNA differ in their geometry and dimensions.
B-DNA:
DNA assumes the so called B-confirmation indicated by their X-ray diffraction patterns, when it is isolated using counter ions which are alkali metals such as Na+ and the relative humidity is >92%. B-DNA is regarded as the native confirmation (biologically functional) form of DNA and the same can be noticed from the x-ray diffraction pattern of the DNA in intact sperm heads. B-DNA consist of two polynucleotide chains that wind about a common axis with a right handed twist to form a 20°A diameter helix.
Two strands are antiparallel and wrap around each other such that they cannot be separated without unwinding the helix and such type of coiling is known as Pleotropic coiling. The planes of the bases are nearly perpendicular to the helix axis. Two bases forms H-bonds and make planar base pairing. The ideal B-DNA helix has 10bp per turn (360°) a helical twist of 36° per base pair and since the aromatic bases have vanderwaals thickness of 3.4A° (0.34nm) and are particularly stacked on each other bases (base stacking). A, T and G, C base pairs are interchangeable. B-DNA has two deep exterior grooves that wind between its sugars phosphate chains. Grooves are formed as consequence of geometry of base pairs. The angle at which the two sugars protrude from each base pair the angle between two glycosidic bond is 120° and 240° for wide range. As a consequence, more base pairs stack on top of each other. The narrow angle between the sugars on one edge of the base pair generate minor groove and angle on other edge gives major groove.
IF the sugar points away in straight line i.e. at an angle of 180° then the two grooves would be in equal diameter. Asymmetry of deoxyriobose sugar and the nucleotides are responsible for generation of grooves in DNA helix.
Double helical DNA and RNA assume several distinct structures which vary with factors such as the humidity, the identity of the cations used and also with base sequences.
A-DNA:
When the relative humidity is decreased to 75%. B-DNA undergoes a reversible conformational change, so called A- form. X-ray studies indicate that A-DNA forms a wider and flutter right handed helix than B-DNA, with 11 bp/ turn, a rise per base pair of 2.6 A° and pitch of the helix being 28.6 A°. In A-DNA the plane of the bases is tilted 20° with respect to the helix axis, since its helix passes above the major groove side of the base pairs rather than through them as in B-DNA. The major deep and minor groove is shallow. Most self-complimentary oligonucleotides of <10bp crystallized is the A-DNA confirmation. Like B-DNA these molecules exhibit considerable sequence specific confirmation variation. A-DNA has so far been observed in only three biological contexts.
- At the cleavage centre of topoisomerase II
- At the active site of DNA polymerase
Z-DNA: (Left handed helix)
The crystal structure determination of the self-complementary hexa nucleotides by Andrew Wang and Alexander Rich surprisingly revealed a left handed double helix. It has 12 Watson crick base pairs per turn, with a pitch of 44A° and in contrast to A-DNA a deep minor groove and no recognizable major groove.
The base pairs in Z-DNA are flipped 180° relative to those in B-DNA, the repeating unit of Z-DNA is dinucleotide. The line joining successive phosphorus atoms on a polynucleotide structure of Z-DNA, therefore follows a zig- zag path around the helix rather than a smooth curve.
X-ray diffraction studies and NMR studies have shown that complementary polynucleotides with alternating purines and pyrimidine such as poly d (GC) and poly d(GC) and poly d (AC), poly d(GT) take up Z-DNA confirmation at high salt concentration. High salt concentration stabilizes electric repulsion between closet approaching phosphate groups on opposite strands.
 Methylation of cytosine at C5 position also promotes Z-DNA formation as the hydrophobic CH3 at this position is less exposed to solvent in Z-DNA than in B-DNA. Reversible conversion of specific segments of B-DNA to Z-DNA under appropriate circumstances acts as a monitoring mechanism in regulating gene expression. There are indicators that it transiently forms behind actively transcribing RNA polymerase but it has been difficult to prove the in vivo existence of Z-DNA due to lack of particular probes for detecting Z-DNA such as Z-DNA specific antibody.
Methylation of cytosine at C5 position also promotes Z-DNA formation as the hydrophobic CH3 at this position is less exposed to solvent in Z-DNA than in B-DNA. Reversible conversion of specific segments of B-DNA to Z-DNA under appropriate circumstances acts as a monitoring mechanism in regulating gene expression. There are indicators that it transiently forms behind actively transcribing RNA polymerase but it has been difficult to prove the in vivo existence of Z-DNA due to lack of particular probes for detecting Z-DNA such as Z-DNA specific antibody.| Parameter | B-Form | A-form | Z-form |
| Helix handedness | Right | Right | Left |
| Base pairs per turn (helical repeat) | 10.5 | 11 | 12 |
| Base pairs per repeating unit | 1 | 1 | 2 |
| Rise per base pair (A0) | 3.4 | 2.6 | 3.7 |
| Pitch/turn of helix | 24.6 Å | 33.2 Å | 45.6 Å |
| Mean propeller twist | +18° | +16° | 0° |
| Glycosyl angle | anti | Anti | C: anti, |
| Sugar pucker | C3'-endo | C2'-endo | C: C2'-endo, |
| Diameter | 25.5 Å | 23.7 Å | 18.4 Å |
| Minor groove width (A0) | 5.7 | 11.0 | 2.0 |
| Minor groove depth (A0) | 7.5 | 2.8 | 13.8 |
Published date : 19 Jun 2014 03:36PM










