Transposons
Sakshi Education
Transposable Elements (TEs), or Transposons, commonly known as "jumping genes", were first discovered by Barbara McClintock. Transposons are defined as “specific DNA segments that can repeatedly insert into one or more sites in one or more genomes”. Transposons are found in almost all organisms and are ubiquitous components of prokaryote and eukaryote genomes. In prokaryotes they transpose to/from cell’s chromosome, plasmid, or a phage chromosome.
In Eukaryotes they transpose to/from same or a different chromosome. Transposons are typically present in large numbers constituting approximately 50% of the human genome and up to 90% of the maize genome. Basically Transposons are classified into two categories of: DNA Transposons and Retrotransposons
DNA Transposons or Class II elements:
DNA Transposons in Prokaryotes are of two types – IS elements (Insertion sequences), Tn elements (Transposons).
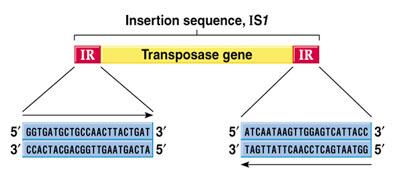
Insertion sequence (IS) elements
IS elements are the simplest type of transposable element found in bacterial chromosomes and plasmids range in size from 768 bp to 5 kb. They encode gene (transposase) for mobilization and insertion. All known IS elements have inverted terminal repeats (ITRs) at their ends. First IS element- IS1 was identified in E. coli’s glactose operon having a size of 768 bp long, 4-19 copies in the E. coli chromosome.
Transposons (Tn):
.jpg)
These elements are similar to IS elements but are more complex structurally and carry additional genes and are of two types: Composite transposons and Noncomposite Transposons.
Composite transposons (Tn) carry genes (example might be a gene for antibiotic resistance) flanked on both sides by IS elements. For example, Tn10 is 9.3 kb and includes 6.5 kb of central DNA (includes a gene for tetracycline resistance) and 1.4 kb inverted IS elements. IS elements supply transposase and ITR recognition signals.
Noncomposite transposons (Tn) also carry genes (example might be a gene for antibiotic resistance) but do not terminate with IS elements. They posses ends that are non-IS element repeated sequences. For example Tn3 is 5 kb with 38-bp ITRs and includes 3 genes; bla (b-lactamase), tnpA (transposase), and tnpB (resolvase, which functions in recombination).
Eukaryotic DNA transposons can be split into four classes: ‘cut and paste’, replicative, RC (Helitrons) and self-synthesizing transposons (Polintons).
‘Cut and paste’ DNA transposons: transposons that belong to this category are excised (or ‘cut’) from their original place as dsDNAs and inserted (or ‘pasted’) into another genomic site. Only a transposon transposed from a recently replicated region of the host genome into a region about to undergo replication can multiply by this mechanism. Both excision and DNA transfer reactions are catalyzed by the transposase enzyme, encoded by the autonomous transposons.
Eg: Ac-Ds element (Activator and Deactivating sequences) in Maize.
P (Penelope) elements in Drosophila.(P elements are responsible for Hybrid Dysgenesis),
Replicative DNA transposons: this category includes any DNA transposon which is not excised during its transposition but rather serves as a template for synthesis of its own copy, which is then inserted into some other genomic site. The synthesis relies on the host repair system. Thus far, the only known super family of eukaryotic DNA transposons using replicative transposition mechanism are MuDR transposons, which function as replicative transposons in germ-line cells and as ‘cut and paste’ transposons in somatic cells. Both excision and DNA transfer reactions are catalyzed by transposases encoded by autonomous transposons.
Helitrons or Rolling-circle (RC) DNA transposons: transposition of these elements is thought to follow a semi-replicative transposition model, according to which, only one strand of the transposed transposon is transferred from one genomic site to another, where it serves as a template for DNA synthesis catalyzed by the host repair machinery. Endonucleolytic cleavage and DNA transfer reactions necessary for RC transposition are catalyzed by the Rep domain of a protein, encoded by autonomous transposons. In all eukaryotic RC Transposons (Helitrons), the Rep domain is fused with an ATP-dependent DNA Hel I. Helitrons lack (TIR) Terminal inverted repeats. Eg: AthE1 and Atrep from Arabidopsis.
Polintons or Self-synthesizing DNA transposons: a recently discovered class of eukaryotic transposons (Polintons) whose transposition is coupled with their own DNA synthesis, catalyzed by the Polinton-encoded DNA polymerase . Excision and integration of Polintons are catalyzed by ‘cut and paste’-like DNA transposases similar to retroviral integrases and some DNA transposases. Polintons are very long,15-20 Kb and code for multiple proteins unlike other transposons.
Retrotransposons or Class I elements: Retrotransposons require the aid of RNA intermediaries for transposition. First they get transcribed to produce RNA transcripts which further get reverse transcribed into DNA by the action of Reverse transcriptase enzyme and finally get inserted into the target site. They do not encode transposase. They follow a "copy and paste" mechanism where the original copy remains intact and a new copy generated is inserted into a new location.
These Retroposons are further classified into 2 classes:
are characterized by the presence of long terminal repeats (LTRs) on both ends for instance gypsy, copia (Drosophila) and Ty elements (yeast).
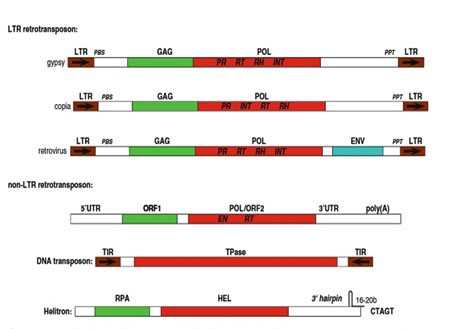
Non-LTR retrotransposons which lack the repeats include both the LINE1or L1, and Alu genes represent families of non-LTR retrotransposons. Depending on the size, these elements are further categorized as Short Interspersed Nuclear Element (SINE) and Long Interspersed Nuclear Element (LINE). L1 (LINE1) elements average about 6 kilobases in length. Alu elements (SINE) average only a few hundred nucleotides. Alu is present as 1 million copies per cell in humans. L1 is also common in humans but present in relatively lesser copies due to its larger size and constitutes up to an estimated 15%–17% of the human genome.
E. Kejnovsky et al. Plant Genome Diversity 2012; 1:17-34
Autonomous and Nonautonomous Transposons
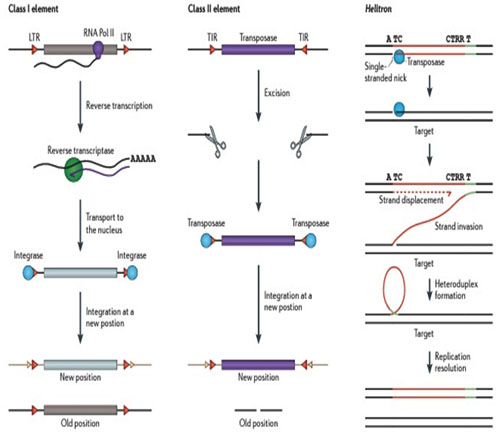
Both DNA transposons and Retrotransposons can be either autonomous or nonautonomous. Autonomous elements as mentioned earlier are self transposing while nonautonomous elements depend on other transposons due to the absence of transposase or reverse transcriptase that is essentialfor theirTransposition.So they rely on other element for the proteins, in order to move.
Ac elements, for example, are autonomous because they can move on their own, whereas Ds elements are nonautonomous because they require the presence of Ac in order to transpose.
Damon Lisch. Nature Reviews Genetics 14, 49-61.
Model of the Polinton transposition.
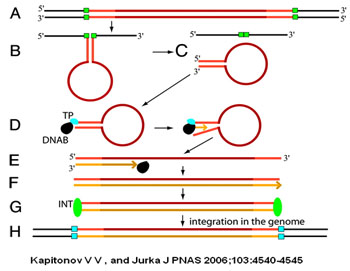
Polinton single-stranded DNAs are shown in red (those synthesized de novo are shown in orange); their TIRs are in light red and orange. The polymerase, terminal protein, and integrase are depicted as black, blue, and green ovals. Old and new target site duplications are marked by small green and blue rectangles.
Biological significance:
Transposons cause genome rearrangements moving from one location to the other. However Transposons lack origins for initiating replication and thus differ from plasmids and bacteriophages which also cause such rearrangements in prokaryotes. The peculiar feature of transposition is that it does not rely on sequence similarity between the donor and recipient sites and thus exhibits random integrations Transposons are not involved in Horizontal gene transfer (moving from one genome to the other)..
Transposons are the major sources of mutations within the genome causing the insertions, deletions and inversions which can occur through direct transposition or reciprocal recombination. Transposons cause gene inactivation by landing inside a gene leading to diseases. For example, insertions of L1 into the factor VIII gene causes hemophilia.
On the other hand, transposons act as a constant force for driving the evolution of genomes by facilitating the translocation of genomic sequences, the shuffling of exons, and the repair of double-stranded breaks. Insertions and transposition can also alter gene regulatory regions and result in phenotypes beneficial to the organism as in case of medaka fish, for instance, the Tol2 DNA transposon is directly linked to pigmentation.
Applications
Transposons act as a source for genes conferring antibiotic genes which are used as selection markers in vectors.
Due to the ability of Transposons to integrate randomly and cause insertional gene inactivation, they are employed as agents to introduce loss of function mutations for identifying functional role of genes by Transposon tagging.
References:
In Eukaryotes they transpose to/from same or a different chromosome. Transposons are typically present in large numbers constituting approximately 50% of the human genome and up to 90% of the maize genome. Basically Transposons are classified into two categories of: DNA Transposons and Retrotransposons
DNA Transposons or Class II elements:
DNA Transposons in Prokaryotes are of two types – IS elements (Insertion sequences), Tn elements (Transposons).
Insertion sequence (IS) elements
IS elements are the simplest type of transposable element found in bacterial chromosomes and plasmids range in size from 768 bp to 5 kb. They encode gene (transposase) for mobilization and insertion. All known IS elements have inverted terminal repeats (ITRs) at their ends. First IS element- IS1 was identified in E. coli’s glactose operon having a size of 768 bp long, 4-19 copies in the E. coli chromosome.
Transposons (Tn):
.jpg)
These elements are similar to IS elements but are more complex structurally and carry additional genes and are of two types: Composite transposons and Noncomposite Transposons.
Composite transposons (Tn) carry genes (example might be a gene for antibiotic resistance) flanked on both sides by IS elements. For example, Tn10 is 9.3 kb and includes 6.5 kb of central DNA (includes a gene for tetracycline resistance) and 1.4 kb inverted IS elements. IS elements supply transposase and ITR recognition signals.
Noncomposite transposons (Tn) also carry genes (example might be a gene for antibiotic resistance) but do not terminate with IS elements. They posses ends that are non-IS element repeated sequences. For example Tn3 is 5 kb with 38-bp ITRs and includes 3 genes; bla (b-lactamase), tnpA (transposase), and tnpB (resolvase, which functions in recombination).
Eukaryotic DNA transposons can be split into four classes: ‘cut and paste’, replicative, RC (Helitrons) and self-synthesizing transposons (Polintons).
‘Cut and paste’ DNA transposons: transposons that belong to this category are excised (or ‘cut’) from their original place as dsDNAs and inserted (or ‘pasted’) into another genomic site. Only a transposon transposed from a recently replicated region of the host genome into a region about to undergo replication can multiply by this mechanism. Both excision and DNA transfer reactions are catalyzed by the transposase enzyme, encoded by the autonomous transposons.
Eg: Ac-Ds element (Activator and Deactivating sequences) in Maize.
P (Penelope) elements in Drosophila.(P elements are responsible for Hybrid Dysgenesis),
Replicative DNA transposons: this category includes any DNA transposon which is not excised during its transposition but rather serves as a template for synthesis of its own copy, which is then inserted into some other genomic site. The synthesis relies on the host repair system. Thus far, the only known super family of eukaryotic DNA transposons using replicative transposition mechanism are MuDR transposons, which function as replicative transposons in germ-line cells and as ‘cut and paste’ transposons in somatic cells. Both excision and DNA transfer reactions are catalyzed by transposases encoded by autonomous transposons.
Helitrons or Rolling-circle (RC) DNA transposons: transposition of these elements is thought to follow a semi-replicative transposition model, according to which, only one strand of the transposed transposon is transferred from one genomic site to another, where it serves as a template for DNA synthesis catalyzed by the host repair machinery. Endonucleolytic cleavage and DNA transfer reactions necessary for RC transposition are catalyzed by the Rep domain of a protein, encoded by autonomous transposons. In all eukaryotic RC Transposons (Helitrons), the Rep domain is fused with an ATP-dependent DNA Hel I. Helitrons lack (TIR) Terminal inverted repeats. Eg: AthE1 and Atrep from Arabidopsis.
Polintons or Self-synthesizing DNA transposons: a recently discovered class of eukaryotic transposons (Polintons) whose transposition is coupled with their own DNA synthesis, catalyzed by the Polinton-encoded DNA polymerase . Excision and integration of Polintons are catalyzed by ‘cut and paste’-like DNA transposases similar to retroviral integrases and some DNA transposases. Polintons are very long,15-20 Kb and code for multiple proteins unlike other transposons.
Retrotransposons or Class I elements: Retrotransposons require the aid of RNA intermediaries for transposition. First they get transcribed to produce RNA transcripts which further get reverse transcribed into DNA by the action of Reverse transcriptase enzyme and finally get inserted into the target site. They do not encode transposase. They follow a "copy and paste" mechanism where the original copy remains intact and a new copy generated is inserted into a new location.
These Retroposons are further classified into 2 classes:
are characterized by the presence of long terminal repeats (LTRs) on both ends for instance gypsy, copia (Drosophila) and Ty elements (yeast).

Non-LTR retrotransposons which lack the repeats include both the LINE1or L1, and Alu genes represent families of non-LTR retrotransposons. Depending on the size, these elements are further categorized as Short Interspersed Nuclear Element (SINE) and Long Interspersed Nuclear Element (LINE). L1 (LINE1) elements average about 6 kilobases in length. Alu elements (SINE) average only a few hundred nucleotides. Alu is present as 1 million copies per cell in humans. L1 is also common in humans but present in relatively lesser copies due to its larger size and constitutes up to an estimated 15%–17% of the human genome.
E. Kejnovsky et al. Plant Genome Diversity 2012; 1:17-34
Autonomous and Nonautonomous Transposons

Both DNA transposons and Retrotransposons can be either autonomous or nonautonomous. Autonomous elements as mentioned earlier are self transposing while nonautonomous elements depend on other transposons due to the absence of transposase or reverse transcriptase that is essentialfor theirTransposition.So they rely on other element for the proteins, in order to move.
Ac elements, for example, are autonomous because they can move on their own, whereas Ds elements are nonautonomous because they require the presence of Ac in order to transpose.
Damon Lisch. Nature Reviews Genetics 14, 49-61.
Model of the Polinton transposition.

Polinton single-stranded DNAs are shown in red (those synthesized de novo are shown in orange); their TIRs are in light red and orange. The polymerase, terminal protein, and integrase are depicted as black, blue, and green ovals. Old and new target site duplications are marked by small green and blue rectangles.
Biological significance:
Transposons cause genome rearrangements moving from one location to the other. However Transposons lack origins for initiating replication and thus differ from plasmids and bacteriophages which also cause such rearrangements in prokaryotes. The peculiar feature of transposition is that it does not rely on sequence similarity between the donor and recipient sites and thus exhibits random integrations Transposons are not involved in Horizontal gene transfer (moving from one genome to the other)..
Transposons are the major sources of mutations within the genome causing the insertions, deletions and inversions which can occur through direct transposition or reciprocal recombination. Transposons cause gene inactivation by landing inside a gene leading to diseases. For example, insertions of L1 into the factor VIII gene causes hemophilia.
On the other hand, transposons act as a constant force for driving the evolution of genomes by facilitating the translocation of genomic sequences, the shuffling of exons, and the repair of double-stranded breaks. Insertions and transposition can also alter gene regulatory regions and result in phenotypes beneficial to the organism as in case of medaka fish, for instance, the Tol2 DNA transposon is directly linked to pigmentation.
Applications
Transposons act as a source for genes conferring antibiotic genes which are used as selection markers in vectors.
Due to the ability of Transposons to integrate randomly and cause insertional gene inactivation, they are employed as agents to introduce loss of function mutations for identifying functional role of genes by Transposon tagging.
References:
- Damon Lisch. (2013). How Important are Transposons for Plant Evolution? Nature Reviews Genetics, 14, 49-61.
- Kapitonov, V. V., and Jurka. J. (2006). Self-synthesizing DNA Transposons in Eukaryotes. PNAS, 103, 4540-4545.
- Kapitonov, V. V., and Jurka. J. (2007). Helitrons on a Roll: Eukaryotic Rolling-circle Transposons. Trends in Genetics, 23(10), 521-529.
Published date : 15 May 2014 02:43PM










