Electron Transport Chain And Oxidative Phosphorylation
Sakshi Education
In aerobic organisms Electron transport chain and Oxidative phosphorylation is the last phase of cellular respiration. All the catabolic pathways (carbohydrate, amino acid, fatty acid) intersect at this stage, where the energy of oxidation drives the synthesis of ATP.
In eukaryotes these reactions takes place in Mitochondria where as in prokaryotes as they lack the cell organelles, electron transport chain and oxidative phosphorylation occurs across the plasma membrane.
Mitochondria:
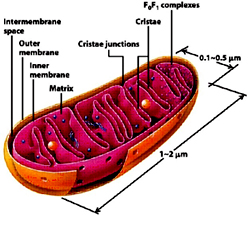
Mitochondrion, a double membrane bound organelle found in the cytoplasm of all eukaryotic cells serves the main function of generating energy in the form of ATP and are hence called - the power house of the cell. Besides it role in generation of ATP it is also involved in metabolism of amino acids and lipids, storage of calcium, cell signaling, generation of heat, cell growth, cell differentiation and death. Mitochondria are typically round to oval in shape ranging in size from 0.5 to 10µm. Unlike other organelles mitochondria is surrounded by a double membrane, possess its own genome and reproduce independent of cell division by binary fission, present in varying numbers per cell. Due to their independent existance from the nuclear DNA and similarities with bacteria, it is believed that mitochondria have originated from bacteria by endosymbiosis.
The mitochondrial shape also varies depending on the cell type, from the tubular-shaped mitochondria in many cell types to mitochondria that consist of sheets and have a spherical-shape structures that are enclosed within two membranes - the outer membrane and the inner membrane. The membranes are made up of phospholipids and proteins. The structures of the various components of mitochondria are as follows:
Steps of protein import into mitochondria General import pathway [Pfanner et.al 1990]:
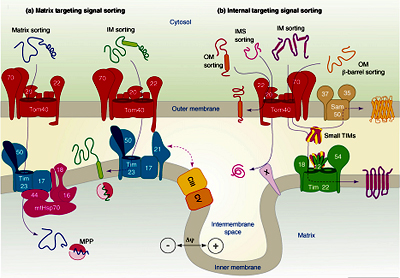 Fig: Protein sorting pathways in Mitochondria.
Fig: Protein sorting pathways in Mitochondria.
Steps involved in oxidative phosphorylation:
 Standard reduction potential of electron carriers
Standard reduction potential of electron carriers
The sequence of electron carriers can be determined by electrode potential and also by using the inhibitors that inhibit the flow of electrons. The order of the carriers: NADH ® Q -® cytochrome b -® cytochrome c1 -® cytochrome c -® cytochrome a --® cytochrome a3-® O2
Electrons are transferred from carriers of lower reduction potential to carriers of higher reduction potential.
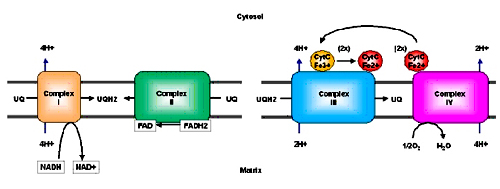
The electron carriers are embedded in the inner mitochondrial membrane as complexes; gentle treatment of inner membrane with detergents reveals the presence of four supramolecular complexes.
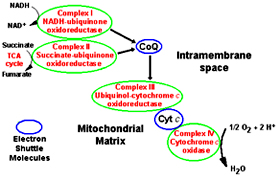 Complex I (NADH-coenzyme Q reductase): catalyze the transfer of electrons from NADH to ubiquinone
Complex I (NADH-coenzyme Q reductase): catalyze the transfer of electrons from NADH to ubiquinone
Complex II (Succinate-coenzyme Q reductase): catalyze the transfer of electrons from FADH2 to ubiquinone
Complex III (Coenzyme Q-cytochrome c reductase): catalyze the transfer of electrons from ubiquinone to cytochrome C
Complex IV (Cytochrome c oxidase): completes the electron transport by catalyzing the transfer of electrons from cytochrome C to oxygen
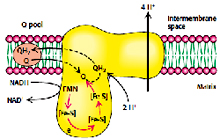
Complex I (NADH-coenzyme Q reductase):
Complex I is a large enzyme complex (>900 kDa) with approximately 46 polypeptide chains, which also contains FMN (flavin mononucleotide) and at least six iron-sulfur (Fe-s) proteins as prosthetic groups. Complex I is L-shaped with one arm lying in the membrane and the other arm projecting in the matrix. This enzyme catalyzes the two coupled processes, a) the exergonic transfer of hydride ion from NADH and proton from matrix to ubiquinone and b) the endergonic transfer of four protons from matrix to intermembrane space.
The overall reaction catalyzed by complex I is:
NADH + ubiquinone + 5 H+ (In) -----® NAD+ + ubiquinol + 4 H+ (Out)
In the first step the electrons are transferred from NADH to FMN to produce FMNH2, this reaction occurs in the matrix side.
NADH + FMN + H+ ® FMNH2 + NAD+
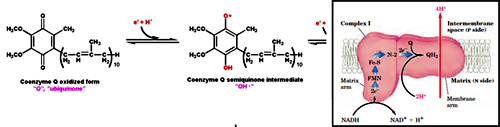 In the second step the electrons are transferred from FMNH2 to a series of Fe-S clusters. FMN can transfer one or two electrons but Fe-S clusters exchanges one electron at a time. Finally the electrons are transferred from Fe-S clusters to Ubiquinone also called as Coenzyme-Q, it has hydrophobic tail which makes it to float in the membrane and allow it to transfer electrons between complex I and Complex III.
In the second step the electrons are transferred from FMNH2 to a series of Fe-S clusters. FMN can transfer one or two electrons but Fe-S clusters exchanges one electron at a time. Finally the electrons are transferred from Fe-S clusters to Ubiquinone also called as Coenzyme-Q, it has hydrophobic tail which makes it to float in the membrane and allow it to transfer electrons between complex I and Complex III.
As the electrons are transferred from NADH (lower reduction potential) to Ubiquinone (higher reduction potential) energy is released which is coupled with the transfer of protons from matrix to inter membrane space. For every NADH, four protons are transferred.
Rotenone, Amytol and Peiricidin A inhibits the transfer of electrons from Fe-S clusters of Complex-I to ubiquinone.
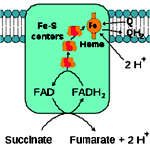
Complex II (Succinate-coenzyme Q reductase):
Complex II is 140 kDa complex which is made of four subunits (polypeptides) and five different prosthetic groups (one FAD, three Fe-S clusters and heme B). Subunits C and D are membrane embedded and Subunits A and B projects towards the matrix (cytosol in case of bacteria). Succinate dehydrogenase which is a enzyme of citiric acid cycle is part of Complex-II, it oxidizes Succinate to fumarate and releasing FADH2. The electrons from FADH2 are transferred to Fe-S clusters and then finally to ubiquinone. Heme B doesn’t take part in electron transport but it prevents the leakage of electrons from the complex there by reducing the formation of reactive oxygen species. Unlike Complex I, protons are not pumped from Complex-II. Acyl CoA dehydrogenase and Glycerol 3 phosphate dehydrogenase also transfers their high energy electrons to FADH2
The reaction catalyzed by Complex-II:
Succinate + Q ® Fumarate+QH2
Malonate and Oxaloacetate inhibits the flow of electrons from complex-II.
Complex III (Coenzyme Q-cytochrome c reductase):
 Complex III also known as cytochrome c reductase or cytochrome bc1 complex is 250kDa complex with 11 polypeptide subunits, an Fe-S cluster (Rieske Fe-S) and three cytochrome (cyt bL, cyt bH and cyt c). Complex III catalyzes the coupled transfer of electrons form ubiquinol to oxidized cytochrome c and vectorial transport of protons to intermembrane space. The net reaction catalyzed by the complex is:
Complex III also known as cytochrome c reductase or cytochrome bc1 complex is 250kDa complex with 11 polypeptide subunits, an Fe-S cluster (Rieske Fe-S) and three cytochrome (cyt bL, cyt bH and cyt c). Complex III catalyzes the coupled transfer of electrons form ubiquinol to oxidized cytochrome c and vectorial transport of protons to intermembrane space. The net reaction catalyzed by the complex is:
QH2 + 2 cyt c2ox + 2H+matrix Q + 2 cyt cred + 4H+ inter membrane space
 Unlike ubiquinone which carries 2 electrons cytochrome c carries only one electron, as only one electron is transferred from QH2 to cytochrome c at a time the reaction mechanism of this complex is complicated and occurs in two steps known as “Q-cycle”.
Unlike ubiquinone which carries 2 electrons cytochrome c carries only one electron, as only one electron is transferred from QH2 to cytochrome c at a time the reaction mechanism of this complex is complicated and occurs in two steps known as “Q-cycle”.
In the first step the three substrates (QH2, cyt c and Q) bind to the complex where QH2 transfers one electron to cyt c and subsequently releasing 2 protons into the intermembrane space, the second electron from QH2 is transferred to Q forming Q2 (ubisemiquinone free radical).
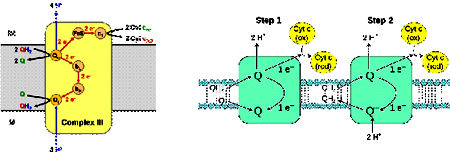 In the second step, second molecule of QH2 binds to the complex and transfer one electron to cyt c as in first step and the second electron is transferred Q- which accepts two protons from the matrix forming QH2. In one Q cycle, two QH2 molecules are oxidized to form two Q molecules, and then one Q molecule is reduced to QH2.
In the second step, second molecule of QH2 binds to the complex and transfer one electron to cyt c as in first step and the second electron is transferred Q- which accepts two protons from the matrix forming QH2. In one Q cycle, two QH2 molecules are oxidized to form two Q molecules, and then one Q molecule is reduced to QH2.
2 QH2 + Q + 2 Cyt cox + 2 H+matrix 2 Q + QH2 + 2 Cyt cred + 4 H+cytoplasm
Antimycin A and British antilewisite (BAL) inhibits the flow of electrons between cytochrome b and c1
Complex IV (Cytochrome c oxidase):
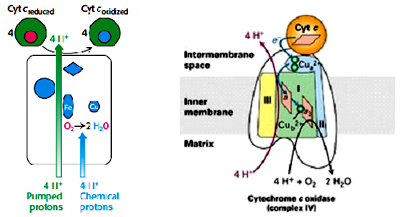 Complex IV is 160 kDa complex with 13 polypeptide subunits, two heme’s (heme a and heme a3) and two copper centers (CuA and CuB). This complex catalyzes the transfer of electrons from reduced cytochrome c to the final electron acceptor oxygen to reduce it to water. Two Cu ions (CuA/CuA) which are complexed with two cysteine residues as in Fe-S clusters forming binuclear center, heme a3 and cuB forms the second binuclear center. Electrons from cytochrome c are transferred to first binuclear center (CuA/CuA), to heme a, to second binuclear center heme a3 and CuB and finally oxygen. For every four electrons passed this complex consumes four protons from the matrix to reduce O2 to H2O, and it also pumps four protons from matrix to inter membrane space. The overall reaction catalyzed by Complex IV is:
Complex IV is 160 kDa complex with 13 polypeptide subunits, two heme’s (heme a and heme a3) and two copper centers (CuA and CuB). This complex catalyzes the transfer of electrons from reduced cytochrome c to the final electron acceptor oxygen to reduce it to water. Two Cu ions (CuA/CuA) which are complexed with two cysteine residues as in Fe-S clusters forming binuclear center, heme a3 and cuB forms the second binuclear center. Electrons from cytochrome c are transferred to first binuclear center (CuA/CuA), to heme a, to second binuclear center heme a3 and CuB and finally oxygen. For every four electrons passed this complex consumes four protons from the matrix to reduce O2 to H2O, and it also pumps four protons from matrix to inter membrane space. The overall reaction catalyzed by Complex IV is:
4 Cyt cred + 8 H+matrix + O2 4 Cyt cox + 2 H2O + 4 H+cytoplasm
Carbon monoxide, azide, hydrogen sulfide and cyanide inhibit cytochrome c oxidase, carbon monoxide reacts with reduced form where as azide reacts with the oxide form of cytochrome.
Overall summary of the flow of electrons and protons through the four complexes of the respiratory chain:
This shuttle functions in Liver, Kidney and Heart mitochondria. The main enzyme involved in this reaction is Malate dehydrogenase; there are two pools of this enzyme mitochondrial and cytosolic malate dehydrogenase.
Electrons from NADH are transferred to Malate in the cytosol forming Oxaloacetate regenerating NAD+ this reaction is catalyzed by cytosolic malate dehydrogenase. The malate formed is transported to matrix by malate-alpha ketoglutarate antiporter which at the same time exports alpha ketoglutarate to cytosol. Malate in the matrix is converted to oxaloacetate and NAD+ present in matrix is reduced to NADH catalyzed by matrix malate dehydrogenase. Since oxaloacetate cannot be transported through the inner membrane, mitochondrial aspartate amino transferase converts oxaloacetate to aspartate which is transported to cytosol through glutamate-aspartate antiporter. In the cytosol, cytosolic aspartate amino transferase converts aspartate to oxaloacetate which again can participate in another cycle.
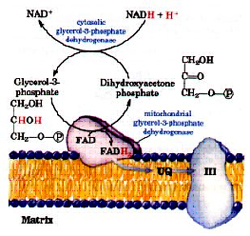
Glycerol-3-phsophate shuttle: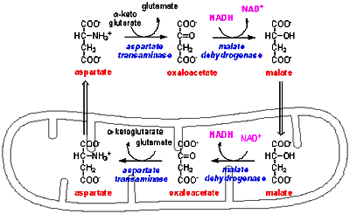
Skeletal muscle and Brain uses another shuttle system to regenerate NADH. A pair of electrons from NADH is transferred to dihydroxyacetone phosphate forming glycerol-3-phosphate regenerating NAD+ catalyzed by cytosolic glycerol-3-phosphate dehydrogenase. Then the electrons from glycerol-3-phosphate are transferred to FAD forming FADH2 and dihydroxyacetone phosphate by the mitochondrial membrane bound glycerol-3-phosphate dehydrogenase.
When Cytosolic NADH is oxidized to NAD+ by malate-aspartate shuttle 2.5 ATP are formed and if it is oxidized by glycerol-3-phosphate shuttle 1.5 ATP are produced.
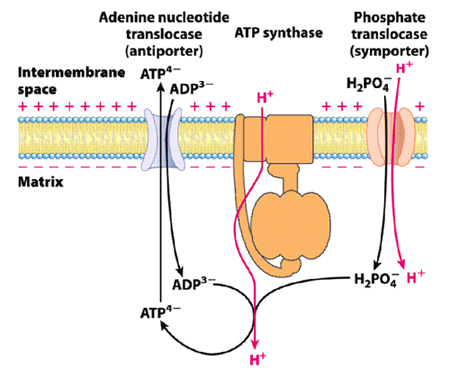
ATP-ADP translocase:
ATP and ADP due to their charge cannot transfer through the mitochondrial inner membrane; a specific translocase, ATP-ADP translocase is involved in the translocation of ATP/ADP through the mitochondrial inner membrane. ATP/ADP is translocated at the expense of 25% (one H+) of the energy yielded from electron transport. ATP/ADP is associated with the translocase without the Mg+2 ions. In actively respiring mitochondria with positive membrane potential ATP export and ADP are import is facilitated. ATP is traversed to the cytosol only when ADP is taken into the matrix and vice versa.
Atractolyside and Bongkrekic acid inhibits ATP-ADP translocase, Atractolyside binds to the cytosolic face whereas Bongkrekic acid binds to the matrix face in either case oxidative phosphorylation is inhibited.
Regulation of Oxidative phosphorylation:
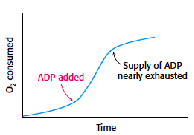
Acceptor control: The level of ADP in a cell determines the rate of oxidative phosphorylation. Electron transport chain and oxidative phosphorylation are coupled; electrons are not transported unless ADP has got phosphorylated to ATP. When the levels of ADP in a cell increases the rate of oxidative phosphorylation increases to meet the requirement of ATP. This regulation of oxidative phosphorylation by ADP is called as acceptor control or respiratory control. In isolated mitochondria when ADP is added externally rate of oxidative phosphorylation increases and it comes back to normal when added ADP is converted to ATP.
Uncoupling:
Certain organism uncouples the electron transport from ATP synthesis to generate heat. Some plants uses uncoupling to evaporate odorant molecules that attract insects towards them to fertilize their flowers.
In animals uncoupling is seen in brown adipose tissue which is involved in non-shivering thermogenesis.
Mitochondrial inner membrane of brown adipose tissue contains a protein called uncoupling protein-1 (UCP-1) or Thermogenin which is similar to ATP-ADP translocase and makes a alternate path for the flow of protons from intermembrane space (cytosol) to matrix without producing ATP. Obesity leads decrease in brown adipose tissue.
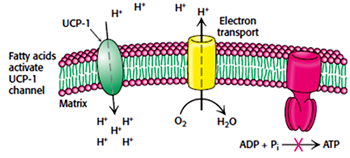
Action of uncoupling protein
Overall when one molecule of Glucose is completely oxidized to water , based on the shuttle system used 32 ATP are formed when malate-aspartate shuttle system is used or 30 ATP when glycerol-3-phosphate shuttle is used.
The number of ATP depends upon the shuttle system used.
In eukaryotes these reactions takes place in Mitochondria where as in prokaryotes as they lack the cell organelles, electron transport chain and oxidative phosphorylation occurs across the plasma membrane.
Mitochondria:
Mitochondrion, a double membrane bound organelle found in the cytoplasm of all eukaryotic cells serves the main function of generating energy in the form of ATP and are hence called - the power house of the cell. Besides it role in generation of ATP it is also involved in metabolism of amino acids and lipids, storage of calcium, cell signaling, generation of heat, cell growth, cell differentiation and death. Mitochondria are typically round to oval in shape ranging in size from 0.5 to 10µm. Unlike other organelles mitochondria is surrounded by a double membrane, possess its own genome and reproduce independent of cell division by binary fission, present in varying numbers per cell. Due to their independent existance from the nuclear DNA and similarities with bacteria, it is believed that mitochondria have originated from bacteria by endosymbiosis.
The mitochondrial shape also varies depending on the cell type, from the tubular-shaped mitochondria in many cell types to mitochondria that consist of sheets and have a spherical-shape structures that are enclosed within two membranes - the outer membrane and the inner membrane. The membranes are made up of phospholipids and proteins. The structures of the various components of mitochondria are as follows:
- Outer membrane
The outer membrane is smooth unlike the inner membrane and has almost the same amount of phospholipids as proteins. It has a large number of special proteins called porin, which allow molecules of 5000 Daltons or less in weight to pass through it. The outer membrane is completely permeable to nutrient molecules, ions, ATP and ADP molecules. Proteins with N-terminal mitochondrial signal sequence binds with TOM Complex (Translocase of outer membrane), which then passes the protein across the membrane. Mitochondria associated ER membrane connects ER (endoplasmic reticulum) to the mitochondria which is important for transfer of lipids and ER-mitochondrial calcium signaling. - Inter membrane space
The space in between outer and inner mitochondrial membrane is called as intermembrane space or perimitochondrial space. The concentration of ions and sugars in intermembrane space is same as that of cytosol due to the freely permeable nature of outer membrane. However the content and type of proteins differs from that of cytosol. Cytochrome c is localized to this space. - Inner Membrane
The inner membrane is more complex in structure than the outer membrane as it contains the complexes of the electron transport chain and the ATP synthetase complex. It is permeable only to oxygen, carbon dioxide and water. It is made up of a large number of proteins that play an important role in producing ATP, and also helps in regulating transfer of metabolites across the membrane. The inner membrane has infoldings called the cristae that increase the surface area for the complexes and proteins that aid in the production of ATP, the energy rich molecules. Inner membrane is rich in phospholipid, Cardiolipin-which makes the membrane impermeable. The TIM Complex (translocase of inner membrane) translocates the ions and molecules across the inner membrane. - Matrix
The matrix is a complex mixture of enzymes that are important for the synthesis of ATP molecules, special mitochondrial ribosome’s, tRNAs and the mitochondrial DNA. Besides these, it has oxygen, carbon dioxide and other recyclable intermediates. Major functions are the oxidation of pyruvate, fatty acids and amino acids.
Steps of protein import into mitochondria General import pathway [Pfanner et.al 1990]:
- Synthesis of proteins on cytosolic ribosome’s as pre-proteins and release into the cytosol.
- Transfer to the mitochondria assisted by cytosolic factors that help to maintain import competence and prevent aggregation.
- Recognition of pre-proteins by interaction of mitochondrial targeting signals with receptors on the surface of the outer membrane of mitochondria
- Initiation of transfer through the pre-protein translocase complex of the outer membrane (TOM complex)
- Interaction of pre-proteins with the surface of the inner membrane and insertion into translocase of the inner membrane (TIM complex) triggered by the membrane potential
- Completion of translocation through outer and inner membrane by the matrix-localized mt-Hsp70-ATP–dependent driving system associated with the TIM complex
- Proteolytic processing of pre-proteins with cleavable targeting signals in the matrix
- Folding of proteins in the matrix assisted by the molecular chaperone systems, mt-Hsp70, Hsp60, and associated co-chaperone.
 Fig: Protein sorting pathways in Mitochondria.
Fig: Protein sorting pathways in Mitochondria.- Pre-sequences direct precursor proteins via the TOM complex to the TIM 23 translocase, where they are sorted to the matrix, inner membrane or intermembrane space. These presequences are of 20-30 amino acids residues capable of folding into a positively charged amphiphilic helix which is important for receptor recognition.
- In comparison, proteins which only contain internal targeting signals are directed to either the inner membrane, intermembrane space or outer membrane, depending on the nature of the signal (Baker MJ et.al 2007).
Steps involved in oxidative phosphorylation:
- Flow of electrons through the membrane bound electron transport chain complexes.
- The transfer of protons to the intermembrane space conserving the energy of oxidation as a transmembrane potential
- Transfer of protons down their concentration gradient through the ATP synthase complex generating the synthesis of ATP.
- Transfer as hydrogen atom
- Transfer as hydride ion and
- Direct transfer of electrons
- Flavoproteins- these participate in one and two electron transfers and they have Fad or FMN as prosthetic groups
- Ubiquinone- also called as coenzyme Q, participates in one and two electron transfers
- Cytochromes- contains heme as prosthetic group, participates in one electron transfer because iron exists in two oxidation states that is either Fe2+ of Fe3+. Five types of cytochromes are involved cyt b, cyt c, cyt c1, cyt a, cyt a3 which differ in their light absorption spectra.
- Iron-sulfur proteins- participate in one electron transfer. Fe-S clusters may be of 2Fe-2S, 4Fe-4S or Rieske Fe-S proteins.
- Copper- which is bounded to protein participates in one electron transfer as it exists either in Cu+ or Cu2+
 Standard reduction potential of electron carriers
Standard reduction potential of electron carriersThe sequence of electron carriers can be determined by electrode potential and also by using the inhibitors that inhibit the flow of electrons. The order of the carriers: NADH ® Q -® cytochrome b -® cytochrome c1 -® cytochrome c -® cytochrome a --® cytochrome a3-® O2
Electrons are transferred from carriers of lower reduction potential to carriers of higher reduction potential.
The electron carriers are embedded in the inner mitochondrial membrane as complexes; gentle treatment of inner membrane with detergents reveals the presence of four supramolecular complexes.
 Complex I (NADH-coenzyme Q reductase): catalyze the transfer of electrons from NADH to ubiquinone
Complex I (NADH-coenzyme Q reductase): catalyze the transfer of electrons from NADH to ubiquinoneComplex II (Succinate-coenzyme Q reductase): catalyze the transfer of electrons from FADH2 to ubiquinone
Complex III (Coenzyme Q-cytochrome c reductase): catalyze the transfer of electrons from ubiquinone to cytochrome C
Complex IV (Cytochrome c oxidase): completes the electron transport by catalyzing the transfer of electrons from cytochrome C to oxygen
Complex I (NADH-coenzyme Q reductase):
Complex I is a large enzyme complex (>900 kDa) with approximately 46 polypeptide chains, which also contains FMN (flavin mononucleotide) and at least six iron-sulfur (Fe-s) proteins as prosthetic groups. Complex I is L-shaped with one arm lying in the membrane and the other arm projecting in the matrix. This enzyme catalyzes the two coupled processes, a) the exergonic transfer of hydride ion from NADH and proton from matrix to ubiquinone and b) the endergonic transfer of four protons from matrix to intermembrane space.
The overall reaction catalyzed by complex I is:
NADH + ubiquinone + 5 H+ (In) -----® NAD+ + ubiquinol + 4 H+ (Out)
In the first step the electrons are transferred from NADH to FMN to produce FMNH2, this reaction occurs in the matrix side.
NADH + FMN + H+ ® FMNH2 + NAD+
 In the second step the electrons are transferred from FMNH2 to a series of Fe-S clusters. FMN can transfer one or two electrons but Fe-S clusters exchanges one electron at a time. Finally the electrons are transferred from Fe-S clusters to Ubiquinone also called as Coenzyme-Q, it has hydrophobic tail which makes it to float in the membrane and allow it to transfer electrons between complex I and Complex III.
In the second step the electrons are transferred from FMNH2 to a series of Fe-S clusters. FMN can transfer one or two electrons but Fe-S clusters exchanges one electron at a time. Finally the electrons are transferred from Fe-S clusters to Ubiquinone also called as Coenzyme-Q, it has hydrophobic tail which makes it to float in the membrane and allow it to transfer electrons between complex I and Complex III.As the electrons are transferred from NADH (lower reduction potential) to Ubiquinone (higher reduction potential) energy is released which is coupled with the transfer of protons from matrix to inter membrane space. For every NADH, four protons are transferred.
Rotenone, Amytol and Peiricidin A inhibits the transfer of electrons from Fe-S clusters of Complex-I to ubiquinone.
Complex II (Succinate-coenzyme Q reductase):
Complex II is 140 kDa complex which is made of four subunits (polypeptides) and five different prosthetic groups (one FAD, three Fe-S clusters and heme B). Subunits C and D are membrane embedded and Subunits A and B projects towards the matrix (cytosol in case of bacteria). Succinate dehydrogenase which is a enzyme of citiric acid cycle is part of Complex-II, it oxidizes Succinate to fumarate and releasing FADH2. The electrons from FADH2 are transferred to Fe-S clusters and then finally to ubiquinone. Heme B doesn’t take part in electron transport but it prevents the leakage of electrons from the complex there by reducing the formation of reactive oxygen species. Unlike Complex I, protons are not pumped from Complex-II. Acyl CoA dehydrogenase and Glycerol 3 phosphate dehydrogenase also transfers their high energy electrons to FADH2
The reaction catalyzed by Complex-II:
Succinate + Q ® Fumarate+QH2
Malonate and Oxaloacetate inhibits the flow of electrons from complex-II.
Complex III (Coenzyme Q-cytochrome c reductase):
 Complex III also known as cytochrome c reductase or cytochrome bc1 complex is 250kDa complex with 11 polypeptide subunits, an Fe-S cluster (Rieske Fe-S) and three cytochrome (cyt bL, cyt bH and cyt c). Complex III catalyzes the coupled transfer of electrons form ubiquinol to oxidized cytochrome c and vectorial transport of protons to intermembrane space. The net reaction catalyzed by the complex is:
Complex III also known as cytochrome c reductase or cytochrome bc1 complex is 250kDa complex with 11 polypeptide subunits, an Fe-S cluster (Rieske Fe-S) and three cytochrome (cyt bL, cyt bH and cyt c). Complex III catalyzes the coupled transfer of electrons form ubiquinol to oxidized cytochrome c and vectorial transport of protons to intermembrane space. The net reaction catalyzed by the complex is:QH2 + 2 cyt c2ox + 2H+matrix Q + 2 cyt cred + 4H+ inter membrane space
 Unlike ubiquinone which carries 2 electrons cytochrome c carries only one electron, as only one electron is transferred from QH2 to cytochrome c at a time the reaction mechanism of this complex is complicated and occurs in two steps known as “Q-cycle”.
Unlike ubiquinone which carries 2 electrons cytochrome c carries only one electron, as only one electron is transferred from QH2 to cytochrome c at a time the reaction mechanism of this complex is complicated and occurs in two steps known as “Q-cycle”.In the first step the three substrates (QH2, cyt c and Q) bind to the complex where QH2 transfers one electron to cyt c and subsequently releasing 2 protons into the intermembrane space, the second electron from QH2 is transferred to Q forming Q2 (ubisemiquinone free radical).
 In the second step, second molecule of QH2 binds to the complex and transfer one electron to cyt c as in first step and the second electron is transferred Q- which accepts two protons from the matrix forming QH2. In one Q cycle, two QH2 molecules are oxidized to form two Q molecules, and then one Q molecule is reduced to QH2.
In the second step, second molecule of QH2 binds to the complex and transfer one electron to cyt c as in first step and the second electron is transferred Q- which accepts two protons from the matrix forming QH2. In one Q cycle, two QH2 molecules are oxidized to form two Q molecules, and then one Q molecule is reduced to QH2.2 QH2 + Q + 2 Cyt cox + 2 H+matrix 2 Q + QH2 + 2 Cyt cred + 4 H+cytoplasm
Antimycin A and British antilewisite (BAL) inhibits the flow of electrons between cytochrome b and c1
Complex IV (Cytochrome c oxidase):
 Complex IV is 160 kDa complex with 13 polypeptide subunits, two heme’s (heme a and heme a3) and two copper centers (CuA and CuB). This complex catalyzes the transfer of electrons from reduced cytochrome c to the final electron acceptor oxygen to reduce it to water. Two Cu ions (CuA/CuA) which are complexed with two cysteine residues as in Fe-S clusters forming binuclear center, heme a3 and cuB forms the second binuclear center. Electrons from cytochrome c are transferred to first binuclear center (CuA/CuA), to heme a, to second binuclear center heme a3 and CuB and finally oxygen. For every four electrons passed this complex consumes four protons from the matrix to reduce O2 to H2O, and it also pumps four protons from matrix to inter membrane space. The overall reaction catalyzed by Complex IV is:
Complex IV is 160 kDa complex with 13 polypeptide subunits, two heme’s (heme a and heme a3) and two copper centers (CuA and CuB). This complex catalyzes the transfer of electrons from reduced cytochrome c to the final electron acceptor oxygen to reduce it to water. Two Cu ions (CuA/CuA) which are complexed with two cysteine residues as in Fe-S clusters forming binuclear center, heme a3 and cuB forms the second binuclear center. Electrons from cytochrome c are transferred to first binuclear center (CuA/CuA), to heme a, to second binuclear center heme a3 and CuB and finally oxygen. For every four electrons passed this complex consumes four protons from the matrix to reduce O2 to H2O, and it also pumps four protons from matrix to inter membrane space. The overall reaction catalyzed by Complex IV is:4 Cyt cred + 8 H+matrix + O2 4 Cyt cox + 2 H2O + 4 H+cytoplasm
Carbon monoxide, azide, hydrogen sulfide and cyanide inhibit cytochrome c oxidase, carbon monoxide reacts with reduced form where as azide reacts with the oxide form of cytochrome.
Overall summary of the flow of electrons and protons through the four complexes of the respiratory chain:

- The proton gradient generated across the inner mitochondrion membrane by complex I (4H+), complex III (4H+) and complex IV (2H+) represents a tremendous amount of stored energy (produced in coupled reactions with the transport of electrons down molecules with increasing reduction potential, ending in oxygen). This potential will be used ultimately to produce ATP.
- This energy is termed electrochemical potential. The inner mitochondrial matrix becomes negatively charged with regard to the cytosol. There is therefore both a pH gradient and an electrical gradient across this membrane (Mitchell's chemiosmotic hypothesis)
- For each NADH oxidized (i.e. 2 electrons ultimately transferred to oxygen) approximately 10H+ are transported across the membrane.
Alternative Oxidase:
Plant mitochondria have an alternative, cyanide resistant oxidase that transfers electrons to oxygen. In addition to optional pathways for the oxidation of NAD(P)H, plant mitochondria contain pathway for the transfer of electrons from ubiquinol to oxygen that by passes cytochrome c oxidase. This nuclear encoded alternative oxidase is found in plants, many algae and some protozoa. Electron flow through the alternative oxidase is insensitive to classic inhibitors of cytochrome c oxidase (cyanide, azide and carbon monoxide) and those complex III (antimycin A, myxothiazole).
The alternative oxidase is associated with a single polypeptide of approximately 32kDa. The alternative oxidase is tightly bound to the inner mitochondrial membrane and diverts electrons from standard ETC at the level of the ubiquinone pool, transferring electrons from ubiquinol to oxygen and generating water as product. When electrons from NADH oxidation flow through the alternative oxidase is lost as heat and cannot be used for ATP synthesis. At least two of the three sites of portion translocation are bypassed, the resulting proton motive force is diminished, and fewer molecules of ATP are synthesized.
The alternative oxidase has one documented role is during the thermogenesis during flowering in a few plants. Cyanide-insensitive O2 uptake is inhibited by SHAM (salicyl hydroxamic acid) and n-propylgallate.
Chemiosmotic theory proposed by Peter Mitchell is a model for the generation of ATP, this theory suggests that the electrochemical gradient generated during the electron transport is utilized for the synthesis of ATP. The transfer of electrons and the generation of ATP are coupled, inhibition of electron transfer inhibits the generation of ATP and vice versa.
The flow of electrons from NADH through the various components of electron transport finally to oxygen is an exergonic process which is used to pump protons from matrix to inter membrane space.
NADH + 1/2O2 + H+ H2O + NAD+ : ?G0’= -220.1 kJ/mol (-52.6 kcal/mol)
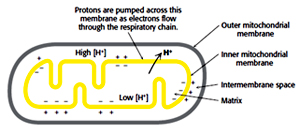 For each pair of electron transferred: 4H+ from complex I, 4H+ from complex III and 2H+ from complex IV are pumped this difference in charge and proton concentration (pH) is stored in the form of proton motive force. Proton motive force has two components (1) the chemical potential energy due to the difference in concentration of a chemical species (H+) in the two regions separated by the membrane, and (2) the electrical potential energy that results from the separation of charge when a proton moves across the membrane without a counter ion.
For each pair of electron transferred: 4H+ from complex I, 4H+ from complex III and 2H+ from complex IV are pumped this difference in charge and proton concentration (pH) is stored in the form of proton motive force. Proton motive force has two components (1) the chemical potential energy due to the difference in concentration of a chemical species (H+) in the two regions separated by the membrane, and (2) the electrical potential energy that results from the separation of charge when a proton moves across the membrane without a counter ion.
Proton motive force (Dp) + chemical gradient (DpH) + charge gradient (DY) The inward flow of protons down their electrochemical gradient drives the synthesis of ATP from ADP and Pi an endergonic process.
ADP + Pi + H+ ATP + H2O: DG0’ = +30.5 kJ/mol (+7.3 kcal/mol)
In addition to the electron carrier proteins embedded in the inner mitochondrial membrane, a special protein ATP synthase is also embedded in the inner membrane also called as complex V , it was originally named as F1F0 ATPase because it was identified to be involved in hydrolysis of ATP. ATP synthase uses the proton gradient created by the electron-transport chain to drive the phosphorylation reaction that generates ATP. ATP synthase is an F-type ATPase which is about 600 kDa and has 16 subunits. It contains two distinct components (ball and stick). The F1 (ball), peripheral membrane subunit and F0 (stick), which is embedded in the membrane. 0-denotes oligomycin sensitive. F1 subunit has the catalytic activity (ATPase).
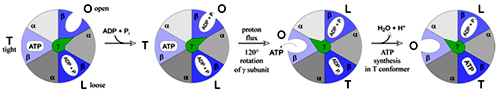 F1 subunit is composed of five polypeptide chains (a3, b3, U, d, and e) a total of nine subunits, a and b subunits forms the hexameric ring (a and b are homologous and are members of P-loop NTPase family) both a and b subunits can bind the nucleotides but the b subunit is involved in catalysis, U subunit forms the helical coil which extends to the center of a3, b3. Nucleotide binding sites of ATP synthase are not equivalent, ? subunit binds to only one of the 3 b subunits, which brings up the conformational change among the three ß subunits, also to their ATP/ADP binding ability and the three conformations are represented by b-ATP, b-ADP and b-empty, these conformations are important for the synthesis of ATP.
F1 subunit is composed of five polypeptide chains (a3, b3, U, d, and e) a total of nine subunits, a and b subunits forms the hexameric ring (a and b are homologous and are members of P-loop NTPase family) both a and b subunits can bind the nucleotides but the b subunit is involved in catalysis, U subunit forms the helical coil which extends to the center of a3, b3. Nucleotide binding sites of ATP synthase are not equivalent, ? subunit binds to only one of the 3 b subunits, which brings up the conformational change among the three ß subunits, also to their ATP/ADP binding ability and the three conformations are represented by b-ATP, b-ADP and b-empty, these conformations are important for the synthesis of ATP.
FO complex forms the proton channel and is composed of three subunits (a, b and c) in the proportion of a, b2, c10-12. Subunit c is hydrophobic polypeptide which is embedded in the membrane, subunit-a binds to the outside of c-ring, two b-subunits, a-subunit and d-subunit are involved in connecting Fo-F1 subunits.
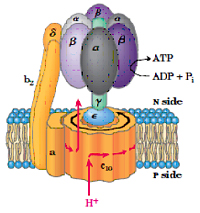
Isotopic labeling experiments suggest that proton gradient is not involved in the generation of ATP but is involved in the release of ATP from the complex. There are three ß subunits in F1 which suggests that there are three active sites in the complex. The proton motive force cause the sequential change in the active sites of ß subunit as protons flow through the complex Various experiments suggest that ATP synthesis is a binding change mechanism, which shows that the ß subunit has three functions. 1) Binding of ADP and Pi, 2) synthesis of ATP and 3) release of ATP. Therefore this subunit is present in three conformations, loose confirmation (L) which binds to ADP and Pi, tight confirmation which brings about the synthesis of ATP and open conformation involved in the release of ATP. Interconversion between these three conformations is driven by ? subunit and each ß subunit changes its conformation from O to T to L with no two conformations present at the single time.
Binding change mechanism of ATP synthesis
The flow of proton through the half channels of a-subunit drives the rotation of c-subunit; the c-subunit is associated ? and d subunits rotates these subunits which in rotates and changes the conformation of a3ß3 subunits and drives the synthesis of ATP.
The number of c-subunits (approx. 10-14) determines the number of protons to be pumped to generate the synthesis of ATP. Each 360o rotation of ?-subunit generates the synthesis of 3 ATP. If there are 10 c-subunits it shows 3.33 protons has to be pumped through c-subunit.
P/O ratio: It denotes the amount of ATP produced from the transport of 2 e- through the electron transport chain. The number of H+ that are transported out for 2 e-is 10 for NADH and 6 for FADH2 and the number of protons that are pumped in for the synthesis of one ATP is 4, out of which one proton is required for the transport of inorganic phosphate (phosphate translocase) into the matrix. Therefore the P/O ratio for NADH is 2.5 (3) and for FADH2 is 1.5 (2).
Inhibitors:
Oligomycin inhibits the flow of protons through the F0 subunit. CCCP and 2, 4-dinitrophenol (ionophores) inhibits ATP synthase by disrupting the membrane potential by carrying the protons across the inner membrane. Oligomycin inhibits cellular respiration whereas 2, 4-DNP inhibits ATP production but increases cellular respiration.
Shuttle systems:
Mitochondrial inner membrane is impermeable to most of the molecules. NADH, generated from glycolysis has to be reoxidized to NAD+ as inner membrane is impermeable to either NADH or NAD+ there should be some mechanism to reoxidize NADH and for the glycolysis to occur continuously.
Shuttle systems play an important role in oxidizing NADH, where by an indirect route electrons from NADH are transported across the inner membrane.
There are two shuttle involved in this process:
Plant mitochondria have an alternative, cyanide resistant oxidase that transfers electrons to oxygen. In addition to optional pathways for the oxidation of NAD(P)H, plant mitochondria contain pathway for the transfer of electrons from ubiquinol to oxygen that by passes cytochrome c oxidase. This nuclear encoded alternative oxidase is found in plants, many algae and some protozoa. Electron flow through the alternative oxidase is insensitive to classic inhibitors of cytochrome c oxidase (cyanide, azide and carbon monoxide) and those complex III (antimycin A, myxothiazole).
The alternative oxidase is associated with a single polypeptide of approximately 32kDa. The alternative oxidase is tightly bound to the inner mitochondrial membrane and diverts electrons from standard ETC at the level of the ubiquinone pool, transferring electrons from ubiquinol to oxygen and generating water as product. When electrons from NADH oxidation flow through the alternative oxidase is lost as heat and cannot be used for ATP synthesis. At least two of the three sites of portion translocation are bypassed, the resulting proton motive force is diminished, and fewer molecules of ATP are synthesized.
The alternative oxidase has one documented role is during the thermogenesis during flowering in a few plants. Cyanide-insensitive O2 uptake is inhibited by SHAM (salicyl hydroxamic acid) and n-propylgallate.
Chemiosmotic theory proposed by Peter Mitchell is a model for the generation of ATP, this theory suggests that the electrochemical gradient generated during the electron transport is utilized for the synthesis of ATP. The transfer of electrons and the generation of ATP are coupled, inhibition of electron transfer inhibits the generation of ATP and vice versa.
The flow of electrons from NADH through the various components of electron transport finally to oxygen is an exergonic process which is used to pump protons from matrix to inter membrane space.
NADH + 1/2O2 + H+ H2O + NAD+ : ?G0’= -220.1 kJ/mol (-52.6 kcal/mol)
 For each pair of electron transferred: 4H+ from complex I, 4H+ from complex III and 2H+ from complex IV are pumped this difference in charge and proton concentration (pH) is stored in the form of proton motive force. Proton motive force has two components (1) the chemical potential energy due to the difference in concentration of a chemical species (H+) in the two regions separated by the membrane, and (2) the electrical potential energy that results from the separation of charge when a proton moves across the membrane without a counter ion.
For each pair of electron transferred: 4H+ from complex I, 4H+ from complex III and 2H+ from complex IV are pumped this difference in charge and proton concentration (pH) is stored in the form of proton motive force. Proton motive force has two components (1) the chemical potential energy due to the difference in concentration of a chemical species (H+) in the two regions separated by the membrane, and (2) the electrical potential energy that results from the separation of charge when a proton moves across the membrane without a counter ion.Proton motive force (Dp) + chemical gradient (DpH) + charge gradient (DY) The inward flow of protons down their electrochemical gradient drives the synthesis of ATP from ADP and Pi an endergonic process.
ADP + Pi + H+ ATP + H2O: DG0’ = +30.5 kJ/mol (+7.3 kcal/mol)
In addition to the electron carrier proteins embedded in the inner mitochondrial membrane, a special protein ATP synthase is also embedded in the inner membrane also called as complex V , it was originally named as F1F0 ATPase because it was identified to be involved in hydrolysis of ATP. ATP synthase uses the proton gradient created by the electron-transport chain to drive the phosphorylation reaction that generates ATP. ATP synthase is an F-type ATPase which is about 600 kDa and has 16 subunits. It contains two distinct components (ball and stick). The F1 (ball), peripheral membrane subunit and F0 (stick), which is embedded in the membrane. 0-denotes oligomycin sensitive. F1 subunit has the catalytic activity (ATPase).
 F1 subunit is composed of five polypeptide chains (a3, b3, U, d, and e) a total of nine subunits, a and b subunits forms the hexameric ring (a and b are homologous and are members of P-loop NTPase family) both a and b subunits can bind the nucleotides but the b subunit is involved in catalysis, U subunit forms the helical coil which extends to the center of a3, b3. Nucleotide binding sites of ATP synthase are not equivalent, ? subunit binds to only one of the 3 b subunits, which brings up the conformational change among the three ß subunits, also to their ATP/ADP binding ability and the three conformations are represented by b-ATP, b-ADP and b-empty, these conformations are important for the synthesis of ATP.
F1 subunit is composed of five polypeptide chains (a3, b3, U, d, and e) a total of nine subunits, a and b subunits forms the hexameric ring (a and b are homologous and are members of P-loop NTPase family) both a and b subunits can bind the nucleotides but the b subunit is involved in catalysis, U subunit forms the helical coil which extends to the center of a3, b3. Nucleotide binding sites of ATP synthase are not equivalent, ? subunit binds to only one of the 3 b subunits, which brings up the conformational change among the three ß subunits, also to their ATP/ADP binding ability and the three conformations are represented by b-ATP, b-ADP and b-empty, these conformations are important for the synthesis of ATP.FO complex forms the proton channel and is composed of three subunits (a, b and c) in the proportion of a, b2, c10-12. Subunit c is hydrophobic polypeptide which is embedded in the membrane, subunit-a binds to the outside of c-ring, two b-subunits, a-subunit and d-subunit are involved in connecting Fo-F1 subunits.

Isotopic labeling experiments suggest that proton gradient is not involved in the generation of ATP but is involved in the release of ATP from the complex. There are three ß subunits in F1 which suggests that there are three active sites in the complex. The proton motive force cause the sequential change in the active sites of ß subunit as protons flow through the complex Various experiments suggest that ATP synthesis is a binding change mechanism, which shows that the ß subunit has three functions. 1) Binding of ADP and Pi, 2) synthesis of ATP and 3) release of ATP. Therefore this subunit is present in three conformations, loose confirmation (L) which binds to ADP and Pi, tight confirmation which brings about the synthesis of ATP and open conformation involved in the release of ATP. Interconversion between these three conformations is driven by ? subunit and each ß subunit changes its conformation from O to T to L with no two conformations present at the single time.
Binding change mechanism of ATP synthesis
The flow of proton through the half channels of a-subunit drives the rotation of c-subunit; the c-subunit is associated ? and d subunits rotates these subunits which in rotates and changes the conformation of a3ß3 subunits and drives the synthesis of ATP.
The number of c-subunits (approx. 10-14) determines the number of protons to be pumped to generate the synthesis of ATP. Each 360o rotation of ?-subunit generates the synthesis of 3 ATP. If there are 10 c-subunits it shows 3.33 protons has to be pumped through c-subunit.
P/O ratio: It denotes the amount of ATP produced from the transport of 2 e- through the electron transport chain. The number of H+ that are transported out for 2 e-is 10 for NADH and 6 for FADH2 and the number of protons that are pumped in for the synthesis of one ATP is 4, out of which one proton is required for the transport of inorganic phosphate (phosphate translocase) into the matrix. Therefore the P/O ratio for NADH is 2.5 (3) and for FADH2 is 1.5 (2).
Inhibitors:
Oligomycin inhibits the flow of protons through the F0 subunit. CCCP and 2, 4-dinitrophenol (ionophores) inhibits ATP synthase by disrupting the membrane potential by carrying the protons across the inner membrane. Oligomycin inhibits cellular respiration whereas 2, 4-DNP inhibits ATP production but increases cellular respiration.
Shuttle systems:
Mitochondrial inner membrane is impermeable to most of the molecules. NADH, generated from glycolysis has to be reoxidized to NAD+ as inner membrane is impermeable to either NADH or NAD+ there should be some mechanism to reoxidize NADH and for the glycolysis to occur continuously.
Shuttle systems play an important role in oxidizing NADH, where by an indirect route electrons from NADH are transported across the inner membrane.
There are two shuttle involved in this process:
- Malate-Aspartate shuttle and
- Glycerol-3-phosphate shuttle.

This shuttle functions in Liver, Kidney and Heart mitochondria. The main enzyme involved in this reaction is Malate dehydrogenase; there are two pools of this enzyme mitochondrial and cytosolic malate dehydrogenase.
Electrons from NADH are transferred to Malate in the cytosol forming Oxaloacetate regenerating NAD+ this reaction is catalyzed by cytosolic malate dehydrogenase. The malate formed is transported to matrix by malate-alpha ketoglutarate antiporter which at the same time exports alpha ketoglutarate to cytosol. Malate in the matrix is converted to oxaloacetate and NAD+ present in matrix is reduced to NADH catalyzed by matrix malate dehydrogenase. Since oxaloacetate cannot be transported through the inner membrane, mitochondrial aspartate amino transferase converts oxaloacetate to aspartate which is transported to cytosol through glutamate-aspartate antiporter. In the cytosol, cytosolic aspartate amino transferase converts aspartate to oxaloacetate which again can participate in another cycle.
Glycerol-3-phsophate shuttle:

Skeletal muscle and Brain uses another shuttle system to regenerate NADH. A pair of electrons from NADH is transferred to dihydroxyacetone phosphate forming glycerol-3-phosphate regenerating NAD+ catalyzed by cytosolic glycerol-3-phosphate dehydrogenase. Then the electrons from glycerol-3-phosphate are transferred to FAD forming FADH2 and dihydroxyacetone phosphate by the mitochondrial membrane bound glycerol-3-phosphate dehydrogenase.
When Cytosolic NADH is oxidized to NAD+ by malate-aspartate shuttle 2.5 ATP are formed and if it is oxidized by glycerol-3-phosphate shuttle 1.5 ATP are produced.
ATP-ADP translocase:

ATP and ADP due to their charge cannot transfer through the mitochondrial inner membrane; a specific translocase, ATP-ADP translocase is involved in the translocation of ATP/ADP through the mitochondrial inner membrane. ATP/ADP is translocated at the expense of 25% (one H+) of the energy yielded from electron transport. ATP/ADP is associated with the translocase without the Mg+2 ions. In actively respiring mitochondria with positive membrane potential ATP export and ADP are import is facilitated. ATP is traversed to the cytosol only when ADP is taken into the matrix and vice versa.
Atractolyside and Bongkrekic acid inhibits ATP-ADP translocase, Atractolyside binds to the cytosolic face whereas Bongkrekic acid binds to the matrix face in either case oxidative phosphorylation is inhibited.
Regulation of Oxidative phosphorylation:

Acceptor control: The level of ADP in a cell determines the rate of oxidative phosphorylation. Electron transport chain and oxidative phosphorylation are coupled; electrons are not transported unless ADP has got phosphorylated to ATP. When the levels of ADP in a cell increases the rate of oxidative phosphorylation increases to meet the requirement of ATP. This regulation of oxidative phosphorylation by ADP is called as acceptor control or respiratory control. In isolated mitochondria when ADP is added externally rate of oxidative phosphorylation increases and it comes back to normal when added ADP is converted to ATP.
Uncoupling:
Certain organism uncouples the electron transport from ATP synthesis to generate heat. Some plants uses uncoupling to evaporate odorant molecules that attract insects towards them to fertilize their flowers.
In animals uncoupling is seen in brown adipose tissue which is involved in non-shivering thermogenesis.
Mitochondrial inner membrane of brown adipose tissue contains a protein called uncoupling protein-1 (UCP-1) or Thermogenin which is similar to ATP-ADP translocase and makes a alternate path for the flow of protons from intermembrane space (cytosol) to matrix without producing ATP. Obesity leads decrease in brown adipose tissue.

Action of uncoupling protein
Overall when one molecule of Glucose is completely oxidized to water , based on the shuttle system used 32 ATP are formed when malate-aspartate shuttle system is used or 30 ATP when glycerol-3-phosphate shuttle is used.
| ATP Yield from complete oxidation of glucose | ||
| Process | Product | ATP |
| Glycolysis | 2 NADH (2.5 or 1.5) | 5 or 3* |
| Oxidation of Pyruvate | 2 NADH (2.5) | 5 |
| Citric Acid cycle | 6 NADH (2.5) | 15 |
| Total yield |
| 32 or 30 |
The number of ATP depends upon the shuttle system used.
Published date : 02 Jul 2014 06:22PM










